+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-14127 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
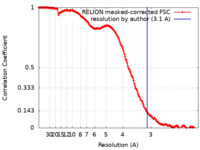
| Title | Bovine complex I in the active state at 3.1 A | |||||||||
 Map data Map data | Consensus map locally sharpened | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationComplex I biogenesis / Mitochondrial protein import / RHOG GTPase cycle / ubiquinone biosynthetic process / Respiratory electron transport / cellular response to oxygen levels / mitochondrial large ribosomal subunit binding / gliogenesis / Neutrophil degranulation / neural precursor cell proliferation ...Complex I biogenesis / Mitochondrial protein import / RHOG GTPase cycle / ubiquinone biosynthetic process / Respiratory electron transport / cellular response to oxygen levels / mitochondrial large ribosomal subunit binding / gliogenesis / Neutrophil degranulation / neural precursor cell proliferation / [2Fe-2S] cluster assembly / oxygen sensor activity / Mitochondrial protein degradation / ubiquinone binding / acyl binding / electron transport coupled proton transport / NADH:ubiquinone reductase (H+-translocating) / acyl carrier activity / mitochondrial ATP synthesis coupled electron transport / NADH dehydrogenase activity / mitochondrial respiratory chain complex I assembly / mitochondrial electron transport, NADH to ubiquinone / respiratory chain complex I / response to cAMP / NADH dehydrogenase (ubiquinone) activity / quinone binding / ATP synthesis coupled electron transport / neurogenesis / reactive oxygen species metabolic process / fatty acid binding / aerobic respiration / respiratory electron transport chain / circadian rhythm / electron transport chain / brain development / mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / mitochondrial membrane / NAD binding / fatty acid biosynthetic process / FMN binding / 4 iron, 4 sulfur cluster binding / oxidoreductase activity / mitochondrial inner membrane / mitochondrial matrix / negative regulation of DNA-templated transcription / apoptotic process / protein-containing complex binding / mitochondrion / nucleoplasm / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Bridges HR / Blaza JN / Yin Z / Chung I / Hirst J | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2023 Journal: Science / Year: 2023Title: Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides. Authors: Hannah R Bridges / James N Blaza / Zhan Yin / Injae Chung / Michael N Pollak / Judy Hirst /   Abstract: The molecular mode of action of biguanides, including the drug metformin, which is widely used in the treatment of diabetes, is incompletely characterized. Here, we define the inhibitory drug-target ...The molecular mode of action of biguanides, including the drug metformin, which is widely used in the treatment of diabetes, is incompletely characterized. Here, we define the inhibitory drug-target interaction(s) of a model biguanide with mammalian respiratory complex I by combining cryo-electron microscopy and enzyme kinetics. We interpret these data to explain the selectivity of biguanide binding to different enzyme states. The primary inhibitory site is in an amphipathic region of the quinone-binding channel, and an additional binding site is in a pocket on the intermembrane-space side of the enzyme. An independent local chaotropic interaction, not previously described for any drug, displaces a portion of a key helix in the membrane domain. Our data provide a structural basis for biguanide action and enable the rational design of medicinal biguanides. #1:  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Cryo-EM structures define ubiquinone-10 binding to mitochondrial complex I and conformational transitions accompanying Q-site occupancy Authors: Chung I / Wright JJ / Bridges HR / Ivanov BS / Biner O / Pereira CS / Arantes GM / Hirst J | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14127.map.gz emd_14127.map.gz | 324.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14127-v30.xml emd-14127-v30.xml emd-14127.xml emd-14127.xml | 68.2 KB 68.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14127_fsc.xml emd_14127_fsc.xml | 15.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_14127.png emd_14127.png | 90.3 KB | ||
| Masks |  emd_14127_msk_1.map emd_14127_msk_1.map | 347.6 MB |  Mask map Mask map | |
| Others |  emd_14127_additional_1.map.gz emd_14127_additional_1.map.gz emd_14127_half_map_1.map.gz emd_14127_half_map_1.map.gz emd_14127_half_map_2.map.gz emd_14127_half_map_2.map.gz | 325.8 MB 280 MB 279.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14127 http://ftp.pdbj.org/pub/emdb/structures/EMD-14127 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14127 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14127 | HTTPS FTP |
-Related structure data
| Related structure data |  7qsdMC  7r41C  7r42C  7r43C  7r44C  7r45C  7r46C  7r47C  7r48C  7r4cC  7r4dC  7r4fC  7r4gC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14127.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14127.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus map locally sharpened | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.056 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
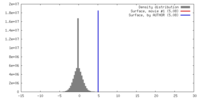
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_14127_msk_1.map emd_14127_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
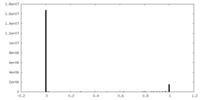
| Density Histograms |
-Additional map: Consensus map globally sharpened
| File | emd_14127_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus map globally sharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
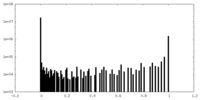
| Density Histograms |
-Half map: Consensus halfmap1
| File | emd_14127_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus halfmap1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
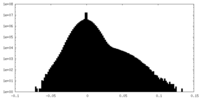
| Density Histograms |
-Half map: Consensus halfmap2
| File | emd_14127_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Consensus halfmap2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : NADH Ubiquinone oxidoreductase (Complex I)
+Supramolecule #1: NADH Ubiquinone oxidoreductase (Complex I)
+Macromolecule #1: NADH-ubiquinone oxidoreductase chain 3
+Macromolecule #2: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial
+Macromolecule #3: NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial
+Macromolecule #4: NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial
+Macromolecule #5: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial
+Macromolecule #6: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial
+Macromolecule #7: NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial
+Macromolecule #8: NADH-ubiquinone oxidoreductase chain 1
+Macromolecule #9: NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial
+Macromolecule #10: NADH-ubiquinone oxidoreductase chain 6
+Macromolecule #11: NADH-ubiquinone oxidoreductase chain 4L
+Macromolecule #12: NADH-ubiquinone oxidoreductase chain 5
+Macromolecule #13: NADH-ubiquinone oxidoreductase chain 4
+Macromolecule #14: NADH-ubiquinone oxidoreductase chain 2
+Macromolecule #15: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mi...
+Macromolecule #16: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mit...
+Macromolecule #17: NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial
+Macromolecule #18: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial
+Macromolecule #19: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2
+Macromolecule #20: Acyl carrier protein, mitochondrial
+Macromolecule #21: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5
+Macromolecule #22: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6
+Macromolecule #23: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8
+Macromolecule #24: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 11
+Macromolecule #25: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13
+Macromolecule #26: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1
+Macromolecule #27: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 3
+Macromolecule #28: NADH dehydrogenase [ubiquinone] 1 subunit C1, mitochondrial
+Macromolecule #29: NADH dehydrogenase [ubiquinone] 1 subunit C2
+Macromolecule #30: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5
+Macromolecule #31: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1
+Macromolecule #32: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mit...
+Macromolecule #33: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 5, mito...
+Macromolecule #34: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 6
+Macromolecule #35: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 2, mito...
+Macromolecule #36: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3
+Macromolecule #37: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mito...
+Macromolecule #38: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4
+Macromolecule #39: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9
+Macromolecule #40: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7
+Macromolecule #41: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10
+Macromolecule #42: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12
+Macromolecule #43: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7
+Macromolecule #44: NADH dehydrogenase [ubiquinone] flavoprotein 3, mitochondrial
+Macromolecule #45: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+Macromolecule #46: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+Macromolecule #47: IRON/SULFUR CLUSTER
+Macromolecule #48: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #49: FLAVIN MONONUCLEOTIDE
+Macromolecule #50: CARDIOLIPIN
+Macromolecule #51: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #52: GUANOSINE-5'-TRIPHOSPHATE
+Macromolecule #53: MAGNESIUM ION
+Macromolecule #54: NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
+Macromolecule #55: ZINC ION
+Macromolecule #56: ~{S}-[2-[3-[[(2~{R})-3,3-dimethyl-2-oxidanyl-4-phosphonooxy-butan...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.14 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil R0.6/1 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 90 sec. / Pretreatment - Atmosphere: AIR Details: Grid also covalently modified by peg-thiol in a nitrogen atmosphere for 48 hours. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







































































































 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)