+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4040 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of mammalian respiratory Complex I, class1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NADH:ubiquinone oxidoreductase / multienzyme complexes / Complex I / mitochondria / oxidoreductase | |||||||||
| Function / homology |  Function and homology information Function and homology informationComplex I biogenesis / RHOG GTPase cycle / Respiratory electron transport / cellular response to oxygen levels / mitochondrial large ribosomal subunit binding / gliogenesis / Neutrophil degranulation / neural precursor cell proliferation / [2Fe-2S] cluster assembly / oxygen sensor activity ...Complex I biogenesis / RHOG GTPase cycle / Respiratory electron transport / cellular response to oxygen levels / mitochondrial large ribosomal subunit binding / gliogenesis / Neutrophil degranulation / neural precursor cell proliferation / [2Fe-2S] cluster assembly / oxygen sensor activity / Mitochondrial protein degradation / ubiquinone binding / acyl binding / electron transport coupled proton transport / NADH:ubiquinone reductase (H+-translocating) / acyl carrier activity / mitochondrial ATP synthesis coupled electron transport / NADH dehydrogenase activity / mitochondrial respiratory chain complex I assembly / mitochondrial electron transport, NADH to ubiquinone / respiratory chain complex I / NADH dehydrogenase (ubiquinone) activity / quinone binding / ATP synthesis coupled electron transport / neurogenesis / reactive oxygen species metabolic process / fatty acid binding / aerobic respiration / respiratory electron transport chain / mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / mitochondrial membrane / NAD binding / fatty acid biosynthetic process / FMN binding / 4 iron, 4 sulfur cluster binding / oxidoreductase activity / mitochondrial inner membrane / mitochondrial matrix / negative regulation of DNA-templated transcription / apoptotic process / mitochondrion / nucleoplasm / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.27 Å | |||||||||
 Authors Authors | Vinothkumar KR / Zhu J | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2016 Journal: Nature / Year: 2016Title: Structure of mammalian respiratory complex I. Authors: Jiapeng Zhu / Kutti R Vinothkumar / Judy Hirst /  Abstract: Complex I (NADH:ubiquinone oxidoreductase), one of the largest membrane-bound enzymes in the cell, powers ATP synthesis in mammalian mitochondria by using the reducing potential of NADH to drive ...Complex I (NADH:ubiquinone oxidoreductase), one of the largest membrane-bound enzymes in the cell, powers ATP synthesis in mammalian mitochondria by using the reducing potential of NADH to drive protons across the inner mitochondrial membrane. Mammalian complex I (ref. 1) contains 45 subunits, comprising 14 core subunits that house the catalytic machinery (and are conserved from bacteria to humans) and a mammalian-specific cohort of 31 supernumerary subunits. Knowledge of the structures and functions of the supernumerary subunits is fragmentary. Here we describe a 4.2-Å resolution single-particle electron cryomicroscopy structure of complex I from Bos taurus. We have located and modelled all 45 subunits, including the 31 supernumerary subunits, to provide the entire structure of the mammalian complex. Computational sorting of the particles identified different structural classes, related by subtle domain movements, which reveal conformationally dynamic regions and match biochemical descriptions of the 'active-to-de-active' enzyme transition that occurs during hypoxia. Our structures therefore provide a foundation for understanding complex I assembly and the effects of mutations that cause clinically relevant complex I dysfunctions, give insights into the structural and functional roles of the supernumerary subunits and reveal new information on the mechanism and regulation of catalysis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4040.map.gz emd_4040.map.gz | 165.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4040-v30.xml emd-4040-v30.xml emd-4040.xml emd-4040.xml | 77.4 KB 77.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4040_fsc.xml emd_4040_fsc.xml | 12.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_4040.png emd_4040.png | 27.8 KB | ||
| Filedesc metadata |  emd-4040.cif.gz emd-4040.cif.gz | 13.4 KB | ||
| Others |  emd_4040_half_map_1.map.gz emd_4040_half_map_1.map.gz emd_4040_half_map_2.map.gz emd_4040_half_map_2.map.gz | 139.8 MB 139.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4040 http://ftp.pdbj.org/pub/emdb/structures/EMD-4040 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4040 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4040 | HTTPS FTP |
-Related structure data
| Related structure data |  5ldwMC  4032C  4041C  5lc5C  5ldxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4040.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4040.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.33 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
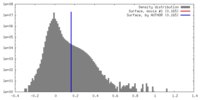
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_4040_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
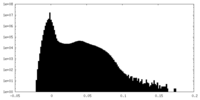
| Density Histograms |
-Half map: #2
| File | emd_4040_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Bovine respiratory Complex I
+Supramolecule #1: Bovine respiratory Complex I
+Macromolecule #1: NADH-ubiquinone oxidoreductase chain 3
+Macromolecule #2: NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial
+Macromolecule #3: NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial
+Macromolecule #4: NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial
+Macromolecule #5: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial
+Macromolecule #6: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial
+Macromolecule #7: NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial,NADH...
+Macromolecule #8: NADH-ubiquinone oxidoreductase chain 1
+Macromolecule #9: NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial
+Macromolecule #10: NADH-ubiquinone oxidoreductase chain 6
+Macromolecule #11: NADH-ubiquinone oxidoreductase chain 4L
+Macromolecule #12: NADH-ubiquinone oxidoreductase chain 5
+Macromolecule #13: NADH-ubiquinone oxidoreductase chain 4
+Macromolecule #14: NADH-ubiquinone oxidoreductase chain 2
+Macromolecule #15: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, ND...
+Macromolecule #16: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, NDUFA9
+Macromolecule #17: NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, NDUFS4
+Macromolecule #18: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, NDUFS6,NAD...
+Macromolecule #19: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2
+Macromolecule #20: Acyl carrier protein, mitochondrial
+Macromolecule #21: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5
+Macromolecule #22: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6
+Macromolecule #23: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8,NADH...
+Macromolecule #24: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 11
+Macromolecule #25: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13, ND...
+Macromolecule #26: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1
+Macromolecule #27: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 3,NADH...
+Macromolecule #28: NADH dehydrogenase [ubiquinone] 1 subunit C1, mitochondrial
+Macromolecule #29: NADH dehydrogenase [ubiquinone] 1 subunit C2, NDUFC2,NADH dehydro...
+Macromolecule #30: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5
+Macromolecule #31: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1
+Macromolecule #32: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mit...
+Macromolecule #33: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 5, mito...
+Macromolecule #34: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 6,NADH ...
+Macromolecule #35: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 2, NDUFB2
+Macromolecule #36: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3, NDUFB3
+Macromolecule #37: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, NDUFB8
+Macromolecule #38: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4, NDUF...
+Macromolecule #39: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9,NADH ...
+Macromolecule #40: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7
+Macromolecule #41: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10, NDU...
+Macromolecule #42: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12
+Macromolecule #43: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7, NDUFA7
+Macromolecule #44: NADH dehydrogenase [ubiquinone] flavoprotein 3, NDUFV3
+Macromolecule #45: IRON/SULFUR CLUSTER
+Macromolecule #46: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #47: FLAVIN MONONUCLEOTIDE
+Macromolecule #48: NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE
+Macromolecule #49: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.5 mg/mL |
|---|---|
| Buffer | pH: 8 / Component - Concentration: 150.0 mM / Component - Formula: NaCl / Component - Name: sodium chloride / Details: 20 mM TRIS-HCL, 150 mM NaCl, 0.03% CYMAL-7 |
| Grid | Model: Quantifoil / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 75 sec. / Pretreatment - Atmosphere: AIR Details: Holey carbon grids were placed on a filter paper in a glass petridish and pumped for around 5 minutes to remove any residual moisture. The glow of the plasma was observed and glow discharge when it was purple. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: HOMEMADE PLUNGER Details: The specimen was vitrified in an environmental plunge-freeze apparatus present in a cold room, blot for 12-15 seconds after the diameter of the blotted meniscus ceases to spread and plunged.. |
| Details | Enzyme was purified from bovine heart mitochondria. The detergent used for the final step is cymal-7. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 87.5 K / Max: 87.5 K |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 1-34 / Average exposure time: 2.0 sec. / Average electron dose: 35.0 e/Å2 Details: Images were collected in movie mode at 17 frames per second. Thus each frame has ~1.0 e/A2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 5.5 µm / Calibrated defocus min: 1.8 µm / Calibrated magnification: 105263 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: OTHER / Overall B value: 83.049 / Target criteria: Maximum likelihood |
|---|---|
| Output model |  PDB-5ldw: |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)