+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-14120 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human RZZ kinetochore corona complex | |||||||||
 Map data Map data | Composite map from focussed refinement maps | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Kinetochore / centromere / chromosome segregation / mitosis / CELL CYCLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein localization to kinetochore involved in kinetochore assembly / Dsl1/NZR complex / RZZ complex / regulation of attachment of spindle microtubules to kinetochore / kinetochore microtubule / centromeric DNA binding / regulation of exit from mitosis / protein localization to kinetochore / retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum / COPI-dependent Golgi-to-ER retrograde traffic ...protein localization to kinetochore involved in kinetochore assembly / Dsl1/NZR complex / RZZ complex / regulation of attachment of spindle microtubules to kinetochore / kinetochore microtubule / centromeric DNA binding / regulation of exit from mitosis / protein localization to kinetochore / retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum / COPI-dependent Golgi-to-ER retrograde traffic / mitotic metaphase chromosome alignment / mitotic spindle assembly checkpoint signaling / Golgi organization / establishment of mitotic spindle orientation / mitotic sister chromatid segregation / endoplasmic reticulum to Golgi vesicle-mediated transport / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / lipid droplet / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / Resolution of Sister Chromatid Cohesion / bioluminescence / generation of precursor metabolites and energy / meiotic cell cycle / RHO GTPases Activate Formins / kinetochore / small GTPase binding / spindle pole / Separation of Sister Chromatids / protein transport / actin cytoskeleton / protein-containing complex assembly / cell division / endoplasmic reticulum membrane / endoplasmic reticulum / nucleus / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
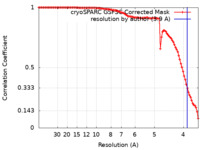
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Raisch T / Ciossani G | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2022 Journal: EMBO J / Year: 2022Title: Structure of the RZZ complex and molecular basis of Spindly-driven corona assembly at human kinetochores. Authors: Tobias Raisch / Giuseppe Ciossani / Ennio d'Amico / Verena Cmentowski / Sara Carmignani / Stefano Maffini / Felipe Merino / Sabine Wohlgemuth / Ingrid R Vetter / Stefan Raunser / Andrea Musacchio /  Abstract: In metazoans, a ≈1 megadalton (MDa) multiprotein complex comprising the dynein-dynactin adaptor Spindly and the ROD-Zwilch-ZW10 (RZZ) complex is the building block of a fibrous biopolymer, the ...In metazoans, a ≈1 megadalton (MDa) multiprotein complex comprising the dynein-dynactin adaptor Spindly and the ROD-Zwilch-ZW10 (RZZ) complex is the building block of a fibrous biopolymer, the kinetochore fibrous corona. The corona assembles on mitotic kinetochores to promote microtubule capture and spindle assembly checkpoint (SAC) signaling. We report here a high-resolution cryo-EM structure that captures the essential features of the RZZ complex, including a farnesyl-binding site required for Spindly binding. Using a highly predictive in vitro assay, we demonstrate that the SAC kinase MPS1 is necessary and sufficient for corona assembly at supercritical concentrations of the RZZ-Spindly (RZZS) complex, and describe the molecular mechanism of phosphorylation-dependent filament nucleation. We identify several structural requirements for RZZS polymerization in rings and sheets. Finally, we identify determinants of kinetochore localization and corona assembly of Spindly. Our results describe a framework for the long-sought-for molecular basis of corona assembly on metazoan kinetochores. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14120.map.gz emd_14120.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14120-v30.xml emd-14120-v30.xml emd-14120.xml emd-14120.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14120_fsc.xml emd_14120_fsc.xml | 9.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_14120.png emd_14120.png | 52.8 KB | ||
| Masks |  emd_14120_msk_1.map emd_14120_msk_1.map emd_14120_msk_2.map emd_14120_msk_2.map | 103 MB 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14120.cif.gz emd-14120.cif.gz | 7.9 KB | ||
| Others |  emd_14120_additional_1.map.gz emd_14120_additional_1.map.gz emd_14120_additional_2.map.gz emd_14120_additional_2.map.gz | 97 MB 2.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14120 http://ftp.pdbj.org/pub/emdb/structures/EMD-14120 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14120 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14120 | HTTPS FTP |
-Validation report
| Summary document |  emd_14120_validation.pdf.gz emd_14120_validation.pdf.gz | 406.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14120_full_validation.pdf.gz emd_14120_full_validation.pdf.gz | 406.4 KB | Display | |
| Data in XML |  emd_14120_validation.xml.gz emd_14120_validation.xml.gz | 11.2 KB | Display | |
| Data in CIF |  emd_14120_validation.cif.gz emd_14120_validation.cif.gz | 14.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14120 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14120 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14120 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14120 | HTTPS FTP |
-Related structure data
| Related structure data |  7qpgMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14120.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14120.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map from focussed refinement maps | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
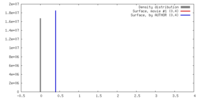
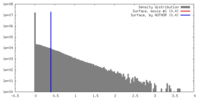
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_14120_msk_1.map emd_14120_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
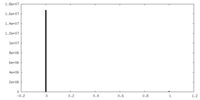
| Density Histograms |
-Mask #2
| File |  emd_14120_msk_2.map emd_14120_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
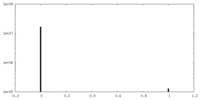
| Density Histograms |
-Additional map: Map from global Non-uniform refinement
| File | emd_14120_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map from global Non-uniform refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Locally filtered map
| File | emd_14120_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Locally filtered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RZZ complex
| Entire | Name: RZZ complex |
|---|---|
| Components |
|
-Supramolecule #1: RZZ complex
| Supramolecule | Name: RZZ complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Protein zwilch homolog
| Macromolecule | Name: Protein zwilch homolog / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 67.293805 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MWERLNCAAE DFYSRLLQKF NEEKKGIRKD PFLYEADVQV QLISKGQPNP LKNILNENDI VFIVEKVPLE KEETSHIEEL QSEETAISD FSTGENVGPL ALPVGKARQL IGLYTMAHNP NMTHLKINLP VTALPPLWVR CDSSDPEGTC WLGAELITTN N SITGIVLY ...String: MWERLNCAAE DFYSRLLQKF NEEKKGIRKD PFLYEADVQV QLISKGQPNP LKNILNENDI VFIVEKVPLE KEETSHIEEL QSEETAISD FSTGENVGPL ALPVGKARQL IGLYTMAHNP NMTHLKINLP VTALPPLWVR CDSSDPEGTC WLGAELITTN N SITGIVLY VVSCKADKNY SVNLENLKNL HKKRHHLSTV TSKGFAQYEL FKSSALDDTI TASQTAIALD ISWSPVDEIL QI PPLSSTA TLNIKVESGE PRGPLNHLYR ELKFLLVLAD GLRTGVTEWL EPLEAKSAVE LVQEFLNDLN KLDGFGDSTK KDT EVETLK HDTAAVDRSV KRLFKVRSDL DFAEQLWCKM SSSVISYQDL VKCFTLIIQS LQRGDIQPWL HSGSNSLLSK LIHQ SYHGT MDTVSLSGTI PVQMLLEIGL DKLKKDYISF FIGQELASLN HLEYFIAPSV DIQEQVYRVQ KLHHILEILV SCMPF IKSQ HELLFSLTQI CIKYYKQNPL DEQHIFQLPV RPTAVKNLYQ SEKPQKWRVE IYSGQKKIKT VWQLSDSSPI DHLNFH KPD FSELTLNGSL EERIFFTNMV TCSQVHFK UniProtKB: Protein zwilch homolog |
-Macromolecule #2: Kinetochore-associated protein 1
| Macromolecule | Name: Kinetochore-associated protein 1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 279.805562 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAHHHHHHSS GVSKGEEDNM AIIKEFMRFK VHMEGSVNGH EFEIEGEGEG RPYEGTQTAK LKVTKGGPLP FAWDILSPQF MYGSKAYVK HPADIPDYLK LSFPEGFKWE RVMNFEDGGV VTVTQDSSLQ DGEFIYKVKL RGTNFPSDGP VMQKKTMGWE A SSERMYPE ...String: MAHHHHHHSS GVSKGEEDNM AIIKEFMRFK VHMEGSVNGH EFEIEGEGEG RPYEGTQTAK LKVTKGGPLP FAWDILSPQF MYGSKAYVK HPADIPDYLK LSFPEGFKWE RVMNFEDGGV VTVTQDSSLQ DGEFIYKVKL RGTNFPSDGP VMQKKTMGWE A SSERMYPE DGALKGEIKQ RLKLKDGGHY DAEVKTTYKA KKPVQLPGAY NVNIKLDITS HNEDYTIVEQ YERAEGRHST GG MDELYKL EVLFQGPGSW NDIELLTNDD TGSGYLSVGS RKEHGTALYQ VDLLVKISSE KASLNPKIQA CSLSDGFIIV ADQ SVILLD SICRSLQLHL VFDTEVDVVG LCQEGKFLLV GERSGNLHLI HVTSKQTLLT NAFVQKANDE NRRTYQNLVI EKDG SNEGT YYMLLLTYSG FFCITNLQLL KIQQAIENVD FSTAKKLQGQ IKSSFISTEN YHTLGCLSLV AGDLASEVPV IIGGT GNCA FSKWEPDSSK KGMTVKNLID AEIIKGAKKF QLIDNLLFVL DTDNVLSLWD IYTLTPVWNW PSLHVEEFLL TTEADS PSS VTWQGITNLK LIALTASANK KMKNLMVYSL PTMEILYSLE VSSVSSLVQT GISTDTIYLL EGVCKNDPKL SEDSVSV LV LRCLTEALPE NRLSRLLHKH RFAEAESFAI QFGLDVELVY KVKSNHILEK LALSSVDASE QTEWQQLVDD AKENLHKI Q DDEFVVNYCL KAQWITYETT QEMLNYAKTR LLKKEDKTAL IYSDGLKEVL RAHAKLTTFY GAFGPEKFSG SSWIEFLNN EDDLKDIFLQ LKEGNLVCAQ YLWLRHRANF ESRFDVKMLE SLLNSMSASV SLQKLCPWFK NDVIPFVRRT VPEGQIILAK WLEQAARNL ELTDKANWPE NGLQLAEIFF TAEKTDELGL ASSWHWISLK DYQNTEEVCQ LRTLVNNLRE LITLHRKYNC K LALSDFEK ENTTTIVFRM FDKVLAPELI PSILEKFIRV YMREHDLQEE ELLLLYIEDL LNRCSSKSTS LFETAWEAKA MA VIACLSD TDLIFDAVLK IMYAAVVPWS AAVEQLVKQH LEMDHPKVKL LQESYKLMEM KKLLRGYGIR EVNLLNKEIM RVV RYILKQ DVPSSLEDAL KVAQAFMLSD DEIYSLRIID LIDREQGEDC LLLLKSLPPA EAEKTAERVI IWARLALQEE PDHS KEGKA WRMSVAKTSV DILKILCDIQ KDNLQKKDEC EEMLKLFKEV ASLQENFEVF LSFEDYSNSS LVADLREQHI KAHEV AQAK HKPGSTPEPI AAEVRSPSME SKLHRQALAL QMSKQELEAE LTLRALKDGN IKTALKKCSD LFKYHCNADT GKLLFL TCQ KLCQMLADNV PVTVPVGLNL PSMIHDLASQ AATICSPDFL LDALELCKHT LMAVELSRQC QMDDCGILMK ASFGTHK DP YEEWSYSDFF SEDGIVLESQ MVLPVIYELI SSLVPLAESK RYPLESTSLP YCSLNEGDGL VLPVINSISA LLQNLQES S QWELALRFVV GSFGTCLQHS VSNFMNATLS EKLFGETTLV KSRHVVMELK EKAVIFIREN ATTLLHKVFN CRLVDLDLA LGYCTLLPQK DVFENLWKLI DKAWQNYDKI LAISLVGSEL ASLYQEIEMG LKFRELSTDA QWGIRLGKLG ISFQPVFRQH FLTKKDLIK ALVENIDMDT SLILEYCSTF QLDCDAVLQL FIETLLHNTN AGQGQGDASM DSAKRRHPKL LAKALEMVPL L TSTKDLVI SLSGILHKLD PYDYEMIEVV LKVIERADEK ITNININQAL SILKHLKSYR RISPPVDLEY QYMLEHVITL PS AAQTRLP FHLIFFGTAQ NFWKILSTEL SEESFPTLLL ISKLMKFSLD TLYVSTAKHV FEKKLKPKLL KLTQAKSSTL INK EITKIT QTIESCLLSI VNPEWAVAIA ISLAQDIPEG SFKISALKFC LYLAERWLQN IPSQDEKREK AEALLKKLHI QYRR SGTEA VLIAHKLNTE EYLRVIGKPA HLIVSLYEHP SINQRIQNSS GTDYPDIHAA AKEIAEVNEI NLEKVWDMLL EKWLC PSTK PGEKPSELFE LQEDEALRRV QYLLLSRPID YSSRMLFVFA TSTTTTLGMH QLTFAHRTRA LQCLFYLADK ETIESL FKK PIEEVKSYLR CITFLASFET LNIPITYELF CSSPKEGMIK GLWKNHSHES MAVRLVTELC LEYKIYDLQL WNGLLQK LL GFNMIPYLRK VLKAISSIHS LWQVPYFSKA WQRVIQIPLL SASCPLSPDQ LSDCSESLIA VLECPVSGDL DLIGVARQ Y IQLELPAFAL ACLMLMPHSE KRHQQIKNFL GSCDPQVILK QLEEHMNTGQ LAGFSHQIRS LILNNIINKK EFGILAKTK YFQMLKMHAM NTNNITELVN YLANDLSLDE ASVLITEYSK HCGKPVPPDT APCEILKMFL SGLS UniProtKB: Uncharacterized protein, Kinetochore-associated protein 1 |
-Macromolecule #3: Centromere/kinetochore protein zw10 homolog
| Macromolecule | Name: Centromere/kinetochore protein zw10 homolog / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 88.940336 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASFVTEVLA HSGRLEKEDL GTRISRLTRR VEEIKGEVCN MISKKYSEFL PSMQSAQGLI TQVDKLSEDI DLLKSRIESE VRRDLHVST GEFTDLKQQL ERDSVVLSLL KQLQEFSTAI EEYNCALTEK KYVTGAQRLE EAQKCLKLLK SRKCFDLKIL K SLSMELTI ...String: MASFVTEVLA HSGRLEKEDL GTRISRLTRR VEEIKGEVCN MISKKYSEFL PSMQSAQGLI TQVDKLSEDI DLLKSRIESE VRRDLHVST GEFTDLKQQL ERDSVVLSLL KQLQEFSTAI EEYNCALTEK KYVTGAQRLE EAQKCLKLLK SRKCFDLKIL K SLSMELTI QKQNILYHLG EEWQKLIVWK FPPSKDTSSL ESYLQTELHL YTEQSHKEEK TPMPPISSVL LAFSVLGELH SK LKSFGQM LLKYILRPLA SCPSLHAVIE SQPNIVIIRF ESIMTNLEYP SPSEVFTKIR LVLEVLQKQL LDLPLDTDLE NEK TSTVPL AEMLGDMIWE DLSECLIKNC LVYSIPTNSS KLQQYEEIIQ STEEFENALK EMRFLKGDTT DLLKYARNIN SHFA NKKCQ DVIVAARNLM TSEIHNTVKI IPDSKINVPE LPTPDEDNKL EVQKVSNTQY HEVMNLEPEN TLDQHSFSLP TCRIS ESVK KLMELAYQTL LEATTSSDQC AVQLFYSVRN IFHLFHDVVP TYHKENLQKL PQLAAIHHNN CMYIAHHLLT LGHQFR LRL APILCDGTAT FVDLVPGFRR LGTECFLAQM RAQKGELLER LSSARNFSNM DDEENYSAAS KAVRQVLHQL KRLGIVW QD VLPVNIYCKA MGTLLNTAIS EVIGKITALE DISTEDGDRL YSLCKTVMDE GPQVFAPLSE ESKNKKYQEE VPVYVPKW M PFKELMMMLQ ASLQEIGDRW ADGKGPLAAA FSSSEVKALI RALFQNTERR AAALAKIK UniProtKB: Centromere/kinetochore protein zw10 homolog |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)