[English] 日本語
 Yorodumi
Yorodumi- EMDB-1391: Structured mRNAs regulate translation initiation by binding to th... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1391 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structured mRNAs regulate translation initiation by binding to the platform of the ribosome. | |||||||||
 Map data Map data | Cryo-EM map of the E.coli 70S pre-initiation complex elucidating the entrapment mechanism by the auto-regulatory S15-rpsOmRNA. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of translation / ribosomal small subunit assembly / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytoplasmic translation / rRNA binding / structural constituent of ribosome / translation / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / negative staining / Resolution: 10.0 Å | |||||||||
 Authors Authors | Marzi S | |||||||||
 Citation Citation |  Journal: Cell / Year: 2007 Journal: Cell / Year: 2007Title: Structured mRNAs regulate translation initiation by binding to the platform of the ribosome. Authors: Stefano Marzi / Alexander G Myasnikov / Alexander Serganov / Chantal Ehresmann / Pascale Romby / Marat Yusupov / Bruno P Klaholz /  Abstract: Gene expression can be regulated at the level of initiation of protein biosynthesis via structural elements present at the 5' untranslated region of mRNAs. These folded mRNA segments may bind to the ...Gene expression can be regulated at the level of initiation of protein biosynthesis via structural elements present at the 5' untranslated region of mRNAs. These folded mRNA segments may bind to the ribosome, thus blocking translation until the mRNA unfolds. Here, we report a series of cryo-electron microscopy snapshots of ribosomal complexes directly visualizing either the mRNA structure blocked by repressor protein S15 or the unfolded, active mRNA. In the stalled state, the folded mRNA prevents the start codon from reaching the peptidyl-tRNA (P) site inside the ribosome. Upon repressor release, the mRNA unfolds and moves into the mRNA channel allowing translation initiation. A comparative structure and sequence analysis suggests the existence of a universal stand-by site on the ribosome (the 30S platform) dedicated for binding regulatory 5' mRNA elements. Different types of mRNA structures may be accommodated during translation preinitiation and regulate gene expression by transiently stalling the ribosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1391.map.gz emd_1391.map.gz | 1.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1391-v30.xml emd-1391-v30.xml emd-1391.xml emd-1391.xml | 11.7 KB 11.7 KB | Display Display |  EMDB header EMDB header |
| Images |  1391.gif 1391.gif | 29 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1391 http://ftp.pdbj.org/pub/emdb/structures/EMD-1391 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1391 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1391 | HTTPS FTP |
-Related structure data
| Related structure data |  2vazMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1391.map.gz / Format: CCP4 / Size: 6.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1391.map.gz / Format: CCP4 / Size: 6.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of the E.coli 70S pre-initiation complex elucidating the entrapment mechanism by the auto-regulatory S15-rpsOmRNA. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
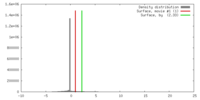
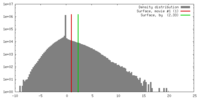
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : 70S pre-initiation complex entrapped by S15-rpsOmRNA
| Entire | Name: 70S pre-initiation complex entrapped by S15-rpsOmRNA |
|---|---|
| Components |
|
-Supramolecule #1000: 70S pre-initiation complex entrapped by S15-rpsOmRNA
| Supramolecule | Name: 70S pre-initiation complex entrapped by S15-rpsOmRNA / type: sample / ID: 1000 Details: 70S E. coli ribosome complexed with S15-rpsOmRNA,stalling Translation Initiation at the pre-initiation step. The structure reveals the molecular details of the Entrapment mechanism. Oligomeric state: monomer / Number unique components: 3 |
|---|---|
| Molecular weight | Theoretical: 2.5 MDa |
-Supramolecule #1: 70S
| Supramolecule | Name: 70S / type: complex / ID: 1 / Name.synonym: 70S / Details: E. coli 70S ribosome / Recombinant expression: No / Ribosome-details: ribosome-prokaryote: ALL |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: rpsO messanger RNA
| Macromolecule | Name: rpsO messanger RNA / type: rna / ID: 1 / Name.synonym: S15mRNA / Classification: OTHER / Structure: SINGLE STRANDED / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: GGGCGAAUUC GAGCUCGGUA CCCAACGUCG CGUAAAUUGU UUAACACUUU GCGUAACGUA CACUGGGAUC GCUGAAUUAG AGAUCGGCGU CCUUUCAUUC UAUAUACUAA GGAGGUUAAA AUGUCUCUAA GUACUGAAGC AACAGCUAAA AUCGUUUCUG AGUUUGGUCG ACCUGCAGGC AUGCA |
-Macromolecule #2: Ribosomal protein S15
| Macromolecule | Name: Ribosomal protein S15 / type: protein_or_peptide / ID: 2 / Name.synonym: S15 / Details: rpsO gene / Oligomeric state: monomeric / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | GO: translation / InterPro: Ribosomal protein S15, bacterial-type |
-Experimental details
-Structure determination
| Method | negative staining, cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 20 mM Tris-HCl pH 7.5, 60 mM KCl, 1mM DTT, 7.5 mM MgCl2 |
| Staining | Type: NEGATIVE / Details: no staining, cryo-EM with holey carbon grids |
| Grid | Details: 300 mesh Cu/Rh |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: home-made cryo-plunger / Method: Blot for 2 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Average: 77 K |
| Alignment procedure | Legacy - Astigmatism: lens astigmatism was corrected at 50,000 times magnification |
| Date | Jan 5, 2005 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: PRIMESCAN / Digitization - Sampling interval: 5 µm / Number real images: 67 / Average electron dose: 20 e/Å2 / Od range: 1.8 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 51484 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Side entry liquid nitrogen-cooled cryo specimen holder Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: individual particles |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 10.0 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: IMAGIC-5 / Details: exact filtered back-projection / Number images used: 31415 |
| Final angle assignment | Details: beta gamma |
-Atomic model buiding 1
| Initial model | (PDB ID:  2aw7  2awb |
|---|---|
| Software | Name: O |
| Details | Protocol: manual. The domains were separately fitted by manual docking using program O |
| Refinement | Protocol: RIGID BODY FIT |
| Output model |  PDB-2vaz: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)