[English] 日本語
 Yorodumi
Yorodumi- EMDB-13828: Single Particle Cryo-EM structure of photosynthetic A10B10 glycer... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single Particle Cryo-EM structure of photosynthetic A10B10 glyceraldehyde-3-phospahte dehydrogenase from Spinacia oleracea. | |||||||||
 Map data Map data | A10B10 photosynthetic glyceraldehyde 3-phosphate dehydrogenase (GAPDH) icosamer. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Photosynthesis / Calvin-Benson cycle / redox regulation / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationglyceraldehyde-3-phosphate dehydrogenase (NADP+) (phosphorylating) / glyceraldehyde-3-phosphate dehydrogenase (NADP+) (phosphorylating) activity / reductive pentose-phosphate cycle / glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity / chloroplast / glucose metabolic process / NAD binding / NADP binding Similarity search - Function | |||||||||
| Biological species |  Spinacia oleracea (spinach) Spinacia oleracea (spinach) | |||||||||
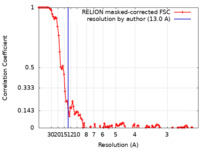
| Method | single particle reconstruction / cryo EM / Resolution: 13.0 Å | |||||||||
 Authors Authors | Marotta R / Fermani S | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Acta Crystallogr D Struct Biol / Year: 2022 Journal: Acta Crystallogr D Struct Biol / Year: 2022Title: Unravelling the regulation pathway of photosynthetic AB-GAPDH. Authors: Roberto Marotta / Alessandra Del Giudice / Libero Gurrieri / Silvia Fanti / Paolo Swuec / Luciano Galantini / Giuseppe Falini / Paolo Trost / Simona Fermani / Francesca Sparla /  Abstract: Oxygenic phototrophs perform carbon fixation through the Calvin-Benson cycle. Different mechanisms adjust the cycle and the light-harvesting reactions to rapid environmental changes. Photosynthetic ...Oxygenic phototrophs perform carbon fixation through the Calvin-Benson cycle. Different mechanisms adjust the cycle and the light-harvesting reactions to rapid environmental changes. Photosynthetic glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is a key enzyme in the cycle. In land plants, different photosynthetic GAPDHs exist: the most abundant isoform is formed by AB heterotetramers and the least abundant by A homotetramers. Regardless of the subunit composition, GAPDH is the major consumer of photosynthetic NADPH and its activity is strictly regulated. While A-GAPDH is regulated by CP12, AB-GAPDH is autonomously regulated through the C-terminal extension (CTE) of its B subunits. Reversible inhibition of AB-GAPDH occurs via the oxidation of a cysteine pair located in the CTE and the substitution of NADP(H) with NAD(H) in the cofactor-binding site. These combined conditions lead to a change in the oligomerization state and enzyme inhibition. SEC-SAXS and single-particle cryo-EM analysis were applied to reveal the structural basis of this regulatory mechanism. Both approaches revealed that spinach (AB)-GAPDH oligomers with n = 1, 2, 4 and 5 co-exist in a dynamic system. B subunits mediate the contacts between adjacent tetramers in AB and AB oligomers. The CTE of each B subunit penetrates into the active site of a B subunit of the adjacent tetramer, which in turn moves its CTE in the opposite direction, effectively preventing the binding of the substrate 1,3-bisphosphoglycerate in the B subunits. The whole mechanism is made possible, and eventually controlled, by pyridine nucleotides. In fact, NAD(H), by removing NADP(H) from A subunits, allows the entrance of the CTE into the active site of the B subunit, hence stabilizing inhibited oligomers. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13828.map.gz emd_13828.map.gz | 14.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13828-v30.xml emd-13828-v30.xml emd-13828.xml emd-13828.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13828_fsc.xml emd_13828_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_13828.png emd_13828.png | 56.5 KB | ||
| Filedesc metadata |  emd-13828.cif.gz emd-13828.cif.gz | 5.8 KB | ||
| Others |  emd_13828_half_map_1.map.gz emd_13828_half_map_1.map.gz emd_13828_half_map_2.map.gz emd_13828_half_map_2.map.gz | 80.9 MB 80.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13828 http://ftp.pdbj.org/pub/emdb/structures/EMD-13828 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13828 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13828 | HTTPS FTP |
-Related structure data
| Related structure data |  7q57MC  7q53C  7q54C  7q55C  7q56C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13828.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13828.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A10B10 photosynthetic glyceraldehyde 3-phosphate dehydrogenase (GAPDH) icosamer. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.21 Å | ||||||||||||||||||||||||||||||||||||
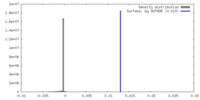
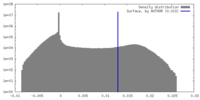
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: A10B10 photosynthetic glyceraldehyde 3-phosphate dehydrogenase (GAPDH) icosamer....
| File | emd_13828_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A10B10 photosynthetic glyceraldehyde 3-phosphate dehydrogenase (GAPDH) icosamer. | ||||||||||||
| Projections & Slices |
| ||||||||||||
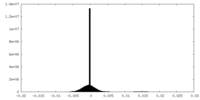
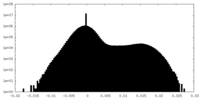
| Density Histograms |
-Half map: A10B10 photosynthetic glyceraldehyde 3-phosphate dehydrogenase (GAPDH) icosamer....
| File | emd_13828_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A10B10 photosynthetic glyceraldehyde 3-phosphate dehydrogenase (GAPDH) icosamer. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : A10B10 glyceraldehyde-3-phospahte dehydrogenase hetero-icosamer c...
| Entire | Name: A10B10 glyceraldehyde-3-phospahte dehydrogenase hetero-icosamer complexed with NAD. |
|---|---|
| Components |
|
-Supramolecule #1: A10B10 glyceraldehyde-3-phospahte dehydrogenase hetero-icosamer c...
| Supramolecule | Name: A10B10 glyceraldehyde-3-phospahte dehydrogenase hetero-icosamer complexed with NAD. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Spinacia oleracea (spinach) Spinacia oleracea (spinach) |
-Macromolecule #1: Glyceraldehyde-3-phosphate dehydrogenase B, chloroplastic
| Macromolecule | Name: Glyceraldehyde-3-phosphate dehydrogenase B, chloroplastic type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO EC number: glyceraldehyde-3-phosphate dehydrogenase (NADP+) (phosphorylating) |
|---|---|
| Source (natural) | Organism:  Spinacia oleracea (spinach) Spinacia oleracea (spinach) |
| Molecular weight | Theoretical: 48.183789 KDa |
| Sequence | String: MASHAALAPS RIPASTRLAS KASQQYSFLT QCSFKRLDVA DFSGLRSSNS VTFTREASFH DVIAAQLTTK PTGAAPVRGE TVAKLKVAI NGFGRIGRNF LRCWHGRKDS PLDVVVVNDS GGVKSATHLL KYDSILGTFK ADVKIIDNET FSIDGKPIKV V SNRDPLKL ...String: MASHAALAPS RIPASTRLAS KASQQYSFLT QCSFKRLDVA DFSGLRSSNS VTFTREASFH DVIAAQLTTK PTGAAPVRGE TVAKLKVAI NGFGRIGRNF LRCWHGRKDS PLDVVVVNDS GGVKSATHLL KYDSILGTFK ADVKIIDNET FSIDGKPIKV V SNRDPLKL PWAELGIDIV IEGTGVFVDG PGAGKHIQAG AKKVIITAPA KGSDIPTYVV GVNEKDYGHD VANIISNASC TT NCLAPFV KVLDEELGIV KGTMTTTHSY TGDQRLLDAS HRDLRRARAA ALNIVPTSTG AAKAVSLVLP QLKGKLNGIA LRV PTPNVS VVDLVVNIEK VGVTAEDVNN AFRKAAAGPL KGVLDVCDIP LVSVDFRCSD FSSTIDSSLT MVMGGDMVKV VAWY DNEWG YSQRVVDLAD LVANKWPGLE GSVASGDPLE DFCKDNPADE ECKLYE UniProtKB: Glyceraldehyde-3-phosphate dehydrogenase B, chloroplastic |
-Macromolecule #2: Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glycera...
| Macromolecule | Name: Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3- ...Name: Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic type: protein_or_peptide / ID: 2 / Number of copies: 10 / Enantiomer: LEVO EC number: glyceraldehyde-3-phosphate dehydrogenase (NADP+) (phosphorylating) |
|---|---|
| Source (natural) | Organism:  Spinacia oleracea (spinach) Spinacia oleracea (spinach) |
| Molecular weight | Theoretical: 36.256391 KDa |
| Sequence | String: KLKVAINGFG RIGRNFLRCW HGRKDSPLDV VVINDTGGVK QASHLLKYDS ILGTFDADVK TAGDSAISVD GKVIKVVSDR NPVNLPWGD MGIDLVIEGT GVFVDRDGAG KHLQAGAKKV LITAPGKGDI PTYVVGVNEE GYTHADTIIS NASCTTNCLA P FVKVLDQK ...String: KLKVAINGFG RIGRNFLRCW HGRKDSPLDV VVINDTGGVK QASHLLKYDS ILGTFDADVK TAGDSAISVD GKVIKVVSDR NPVNLPWGD MGIDLVIEGT GVFVDRDGAG KHLQAGAKKV LITAPGKGDI PTYVVGVNEE GYTHADTIIS NASCTTNCLA P FVKVLDQK FGIIKGTMTT THSYTGDQRL LDASHRDLRR ARAACLNIVP TSTGAAKAVA LVLPNLKGKL NGIALRVPTP NV SVVDLVV QVSKKTFAEE VNAAFRESAD NELKGILSVC DEPLVSIDFR CTDVSSTIDS SLTMVMGDDM VKVIAWYDNE WGY SQRVVD LADIVANKWQ A UniProtKB: Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic |
-Macromolecule #3: NICOTINAMIDE-ADENINE-DINUCLEOTIDE
| Macromolecule | Name: NICOTINAMIDE-ADENINE-DINUCLEOTIDE / type: ligand / ID: 3 / Number of copies: 20 / Formula: NAD |
|---|---|
| Molecular weight | Theoretical: 663.425 Da |
| Chemical component information |  ChemComp-NAD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)