[English] 日本語
 Yorodumi
Yorodumi- EMDB-13824: Single Particle Cryo-EM structure of photosynthetic A2B2 glyceral... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single Particle Cryo-EM structure of photosynthetic A2B2 glyceraldehyde 3-phosphate dehydrogenase from Spinacia oleracia | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Photosynthesis / Calvin-Benson cycle / redox regulation / glyceraldehyde-3-phosphate dehydrogenase / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationglyceraldehyde-3-phosphate dehydrogenase (NADP+) (phosphorylating) / glyceraldehyde-3-phosphate dehydrogenase (NADP+) (phosphorylating) activity / reductive pentose-phosphate cycle / glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity / chloroplast / glucose metabolic process / NAD binding / NADP binding Similarity search - Function | |||||||||
| Biological species |  Spinacia oleracea (spinach) Spinacia oleracea (spinach) | |||||||||
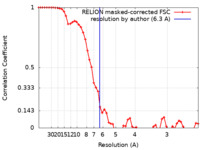
| Method | single particle reconstruction / cryo EM / Resolution: 6.3 Å | |||||||||
 Authors Authors | Marotta R / Fermani S / Sparla F / Trost P / Del Giudice A | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Acta Crystallogr D Struct Biol / Year: 2022 Journal: Acta Crystallogr D Struct Biol / Year: 2022Title: Unravelling the regulation pathway of photosynthetic AB-GAPDH. Authors: Roberto Marotta / Alessandra Del Giudice / Libero Gurrieri / Silvia Fanti / Paolo Swuec / Luciano Galantini / Giuseppe Falini / Paolo Trost / Simona Fermani / Francesca Sparla /  Abstract: Oxygenic phototrophs perform carbon fixation through the Calvin-Benson cycle. Different mechanisms adjust the cycle and the light-harvesting reactions to rapid environmental changes. Photosynthetic ...Oxygenic phototrophs perform carbon fixation through the Calvin-Benson cycle. Different mechanisms adjust the cycle and the light-harvesting reactions to rapid environmental changes. Photosynthetic glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is a key enzyme in the cycle. In land plants, different photosynthetic GAPDHs exist: the most abundant isoform is formed by AB heterotetramers and the least abundant by A homotetramers. Regardless of the subunit composition, GAPDH is the major consumer of photosynthetic NADPH and its activity is strictly regulated. While A-GAPDH is regulated by CP12, AB-GAPDH is autonomously regulated through the C-terminal extension (CTE) of its B subunits. Reversible inhibition of AB-GAPDH occurs via the oxidation of a cysteine pair located in the CTE and the substitution of NADP(H) with NAD(H) in the cofactor-binding site. These combined conditions lead to a change in the oligomerization state and enzyme inhibition. SEC-SAXS and single-particle cryo-EM analysis were applied to reveal the structural basis of this regulatory mechanism. Both approaches revealed that spinach (AB)-GAPDH oligomers with n = 1, 2, 4 and 5 co-exist in a dynamic system. B subunits mediate the contacts between adjacent tetramers in AB and AB oligomers. The CTE of each B subunit penetrates into the active site of a B subunit of the adjacent tetramer, which in turn moves its CTE in the opposite direction, effectively preventing the binding of the substrate 1,3-bisphosphoglycerate in the B subunits. The whole mechanism is made possible, and eventually controlled, by pyridine nucleotides. In fact, NAD(H), by removing NADP(H) from A subunits, allows the entrance of the CTE into the active site of the B subunit, hence stabilizing inhibited oligomers. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13824.map.gz emd_13824.map.gz | 2.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13824-v30.xml emd-13824-v30.xml emd-13824.xml emd-13824.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13824_fsc.xml emd_13824_fsc.xml | 5.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_13824.png emd_13824.png | 66.3 KB | ||
| Filedesc metadata |  emd-13824.cif.gz emd-13824.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13824 http://ftp.pdbj.org/pub/emdb/structures/EMD-13824 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13824 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13824 | HTTPS FTP |
-Related structure data
| Related structure data |  7q53MC  7q54C  7q55C  7q56C  7q57C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13824.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13824.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.21 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Photosynthetic A2B2 glyceraldehyde-3-phosphate dehydrogenase hete...
| Entire | Name: Photosynthetic A2B2 glyceraldehyde-3-phosphate dehydrogenase hetero-tetramer complexed with NAD. |
|---|---|
| Components |
|
-Supramolecule #1: Photosynthetic A2B2 glyceraldehyde-3-phosphate dehydrogenase hete...
| Supramolecule | Name: Photosynthetic A2B2 glyceraldehyde-3-phosphate dehydrogenase hetero-tetramer complexed with NAD. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Spinacia oleracea (spinach) Spinacia oleracea (spinach) |
-Macromolecule #1: Glyceraldehyde-3-phosphate dehydrogenase B, chloroplastic
| Macromolecule | Name: Glyceraldehyde-3-phosphate dehydrogenase B, chloroplastic type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO EC number: glyceraldehyde-3-phosphate dehydrogenase (NADP+) (phosphorylating) |
|---|---|
| Source (natural) | Organism:  Spinacia oleracea (spinach) / Organ: chloroplast Spinacia oleracea (spinach) / Organ: chloroplast |
| Molecular weight | Theoretical: 36.28866 KDa |
| Sequence | String: KLKVAINGFG RIGRNFLRCW HGRKDSPLDV VVVNDSGGVK SATHLLKYDS ILGTFKADVK IIDNETFSID GKPIKVVSNR DPLKLPWAE LGIDIVIEGT GVFVDGPGAG KHIQAGAKKV IITAPAKGSD IPTYVVGVNE KDYGHDVANI ISNASCTTNC L APFVKVLD ...String: KLKVAINGFG RIGRNFLRCW HGRKDSPLDV VVVNDSGGVK SATHLLKYDS ILGTFKADVK IIDNETFSID GKPIKVVSNR DPLKLPWAE LGIDIVIEGT GVFVDGPGAG KHIQAGAKKV IITAPAKGSD IPTYVVGVNE KDYGHDVANI ISNASCTTNC L APFVKVLD EELGIVKGTM TTTHSYTGDQ RLLDASHRDL RRARAAALNI VPTSTGAAKA VSLVLPQLKG KLNGIALRVP TP NVSVVDL VVNIEKVGVT AEDVNNAFRK AAAGPLKGVL DVCDIPLVSV DFRCSDFSST IDSSLTMVMG GDMVKVVAWY DNE WGYSQR VVDLADLVAN KWP UniProtKB: Glyceraldehyde-3-phosphate dehydrogenase B, chloroplastic |
-Macromolecule #2: Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glycera...
| Macromolecule | Name: Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3- ...Name: Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic,Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO EC number: glyceraldehyde-3-phosphate dehydrogenase (NADP+) (phosphorylating) |
|---|---|
| Source (natural) | Organism:  Spinacia oleracea (spinach) / Organ: chloroplast Spinacia oleracea (spinach) / Organ: chloroplast |
| Molecular weight | Theoretical: 36.256391 KDa |
| Sequence | String: KLKVAINGFG RIGRNFLRCW HGRKDSPLDV VVINDTGGVK QASHLLKYDS ILGTFDADVK TAGDSAISVD GKVIKVVSDR NPVNLPWGD MGIDLVIEGT GVFVDRDGAG KHLQAGAKKV LITAPGKGDI PTYVVGVNEE GYTHADTIIS NASCTTNCLA P FVKVLDQK ...String: KLKVAINGFG RIGRNFLRCW HGRKDSPLDV VVINDTGGVK QASHLLKYDS ILGTFDADVK TAGDSAISVD GKVIKVVSDR NPVNLPWGD MGIDLVIEGT GVFVDRDGAG KHLQAGAKKV LITAPGKGDI PTYVVGVNEE GYTHADTIIS NASCTTNCLA P FVKVLDQK FGIIKGTMTT THSYTGDQRL LDASHRDLRR ARAACLNIVP TSTGAAKAVA LVLPNLKGKL NGIALRVPTP NV SVVDLVV QVSKKTFAEE VNAAFRESAD NELKGILSVC DEPLVSIDFR CTDVSSTIDS SLTMVMGDDM VKVIAWYDNE WGY SQRVVD LADIVANKWQ A UniProtKB: Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic |
-Macromolecule #3: NICOTINAMIDE-ADENINE-DINUCLEOTIDE
| Macromolecule | Name: NICOTINAMIDE-ADENINE-DINUCLEOTIDE / type: ligand / ID: 3 / Number of copies: 4 / Formula: NAD |
|---|---|
| Molecular weight | Theoretical: 663.425 Da |
| Chemical component information |  ChemComp-NAD: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)