[English] 日本語
 Yorodumi
Yorodumi- EMDB-13683: Cryo-EM structure of Giardia lamblia ribosome at 2.75 A resolution -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Giardia lamblia ribosome at 2.75 A resolution | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ribosome / giardia / parasite | |||||||||
| Function / homology |  Function and homology information Function and homology informationendonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of LSU-rRNA / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / cytosolic ribosome / ribosomal large subunit biogenesis / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / small-subunit processome / modification-dependent protein catabolic process ...endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of LSU-rRNA / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / cytosolic ribosome / ribosomal large subunit biogenesis / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / small-subunit processome / modification-dependent protein catabolic process / protein tag activity / rRNA processing / ribosome biogenesis / ribosomal small subunit biogenesis / ribosomal small subunit assembly / ribosomal large subunit assembly / 5S rRNA binding / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / negative regulation of translation / rRNA binding / structural constituent of ribosome / protein ubiquitination / ribosome / translation / ribonucleoprotein complex / mRNA binding / ubiquitin protein ligase binding / nucleolus / RNA binding / zinc ion binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Giardia lamblia ATCC 50803 (eukaryote) Giardia lamblia ATCC 50803 (eukaryote) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.75 Å | |||||||||
 Authors Authors | Hiregange DG / Rivalta A / Bose T / Breiner-Goldstein E / Samiya S / Cimicata G / Kulakova L / Zimmerman E / Bashan A / Herzberg O / Yonath A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2022 Journal: Nucleic Acids Res / Year: 2022Title: Cryo-EM structure of the ancient eukaryotic ribosome from the human parasite Giardia lamblia. Authors: Disha-Gajanan Hiregange / Andre Rivalta / Tanaya Bose / Elinor Breiner-Goldstein / Sarit Samiya / Giuseppe Cimicata / Liudmila Kulakova / Ella Zimmerman / Anat Bashan / Osnat Herzberg / Ada Yonath /   Abstract: Giardiasis is a disease caused by the protist Giardia lamblia. As no human vaccines have been approved so far against it, and resistance to current drugs is spreading, new strategies for combating ...Giardiasis is a disease caused by the protist Giardia lamblia. As no human vaccines have been approved so far against it, and resistance to current drugs is spreading, new strategies for combating giardiasis need to be developed. The G. lamblia ribosome may provide a promising therapeutic target due to its distinct sequence differences from ribosomes of most eukaryotes and prokaryotes. Here, we report the cryo-electron microscopy structure of the G. lamblia (WB strain) ribosome determined at 2.75 Å resolution. The ribosomal RNA is the shortest known among eukaryotes, and lacks nearly all the eukaryote-specific ribosomal RNA expansion segments. In contrast, the ribosomal proteins are typically eukaryotic with some species-specific insertions/extensions. Most typical inter-subunit bridges are maintained except for one missing contact site. Unique structural features are located mainly at the ribosome's periphery. These may be exploited as target sites for the design of new compounds that inhibit selectively the parasite's ribosomal activity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13683.map.gz emd_13683.map.gz | 259.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13683-v30.xml emd-13683-v30.xml emd-13683.xml emd-13683.xml | 92.9 KB 92.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13683.png emd_13683.png | 59.7 KB | ||
| Filedesc metadata |  emd-13683.cif.gz emd-13683.cif.gz | 17.5 KB | ||
| Others |  emd_13683_additional_1.map.gz emd_13683_additional_1.map.gz | 59.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13683 http://ftp.pdbj.org/pub/emdb/structures/EMD-13683 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13683 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13683 | HTTPS FTP |
-Related structure data
| Related structure data |  7pwoMC  7pwfC  7pwgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13683.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13683.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_13683_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
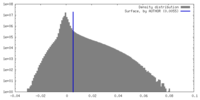
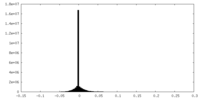
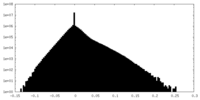
| Density Histograms |
- Sample components
Sample components
+Entire : Giardia lamblia ribosome
+Supramolecule #1: Giardia lamblia ribosome
+Macromolecule #1: Ribosomal protein S19e
+Macromolecule #2: Ribosomal protein S13
+Macromolecule #3: Ribosomal protein S9
+Macromolecule #4: Ribosomal protein S3
+Macromolecule #5: Ribosomal protein S23
+Macromolecule #6: Ribosomal protein S18
+Macromolecule #7: Ribosomal protein S16
+Macromolecule #8: Ribosomal protein S2
+Macromolecule #9: 40S ribosomal protein S3a
+Macromolecule #10: Ribosomal protein S27
+Macromolecule #11: 40S ribosomal protein S26
+Macromolecule #12: 40S ribosomal protein S21
+Macromolecule #13: Ribosomal protein S17
+Macromolecule #14: Ribosomal protein S10B
+Macromolecule #15: 40S ribosomal protein S8
+Macromolecule #16: 40S ribosomal protein S30
+Macromolecule #17: 40S ribosomal protein S7
+Macromolecule #18: Ribosomal protein eL41
+Macromolecule #19: Ribosomal protein S28
+Macromolecule #20: Ribosomal protein S14
+Macromolecule #21: Ribosomal protein S15A
+Macromolecule #22: 40S ribosomal protein S4
+Macromolecule #23: SSU ribosomal protein S17P
+Macromolecule #24: Ribosomal protein S20
+Macromolecule #28: Ribosomal protein L2
+Macromolecule #29: Ribosomal protein L3
+Macromolecule #30: Ribosomal protein L4
+Macromolecule #31: Ribosomal protein L5
+Macromolecule #32: Ribosomal protein L7
+Macromolecule #33: 60S ribosomal protein L7a
+Macromolecule #34: Ribosomal protein L6
+Macromolecule #35: Ribosomal protein L10
+Macromolecule #36: Ribosomal protein L11
+Macromolecule #37: 60S ribosomal protein L13
+Macromolecule #38: Ribosomal protein L14
+Macromolecule #39: Ribosomal protein L15
+Macromolecule #40: Ribosomal protein L13a
+Macromolecule #41: Ribosomal protein L17
+Macromolecule #42: Ribosomal protein L18
+Macromolecule #43: Ribosomal protein L19
+Macromolecule #44: 60S ribosomal protein L18a
+Macromolecule #45: Ribosomal protein L21
+Macromolecule #46: Ribosomal protein eL22
+Macromolecule #47: Ribosomal protein L23
+Macromolecule #48: Ribosomal protein L23A
+Macromolecule #49: Ribosomal protein L26
+Macromolecule #50: 60S ribosomal protein L27
+Macromolecule #51: Ribosomal protein L27a
+Macromolecule #52: 60S ribosomal protein L29
+Macromolecule #53: Ribosomal protein L30
+Macromolecule #54: Ribosomal protein L31B
+Macromolecule #55: Ribosomal protein L32
+Macromolecule #56: Ribosomal protein L35a
+Macromolecule #57: Ribosomal protein L34
+Macromolecule #58: Ribosomal protein L35
+Macromolecule #59: Ribosomal protein L36-1
+Macromolecule #60: Ribosomal protein L37
+Macromolecule #61: Ribosomal L38e
+Macromolecule #62: Ribosomal protein L39
+Macromolecule #63: Ubiquitin/Ribosomal protein L40e
+Macromolecule #64: Ribosomal protein L44
+Macromolecule #65: Ribosomal protein L37a
+Macromolecule #67: Ribosomal protein L24A
+Macromolecule #69: Ribosomal protein S29A
+Macromolecule #70: Ribosomal protein S5
+Macromolecule #71: Ribosomal protein S24
+Macromolecule #72: 40S ribosomal protein S6
+Macromolecule #74: 40S ribosomal protein SA
+Macromolecule #25: rRNA 28S
+Macromolecule #26: rRNA 5S
+Macromolecule #27: rRNA 5.8S
+Macromolecule #66: E-site tRNA
+Macromolecule #68: P-site tRNA
+Macromolecule #73: rRNA 18S
+Macromolecule #75: POTASSIUM ION
+Macromolecule #76: MAGNESIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 5567 / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.75 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION (ver. 3.1) / Number images used: 91058 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)