+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13489 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Scytonema hofmannii TnsC bound to AMPPNP and DNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TnsC / Transposition protein / AAA+ ATPase / Tn7 / CRISPR / DNA BINDING PROTEIN | |||||||||
| Biological species |  Scytonema hofmannii (bacteria) / synthetic construct (others) Scytonema hofmannii (bacteria) / synthetic construct (others) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.57 Å | |||||||||
 Authors Authors | Querques I / Jinek M | |||||||||
| Funding support | European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Target site selection and remodelling by type V CRISPR-transposon systems. Authors: Irma Querques / Michael Schmitz / Seraina Oberli / Christelle Chanez / Martin Jinek /  Abstract: Canonical CRISPR-Cas systems provide adaptive immunity against mobile genetic elements. However, type I-F, I-B and V-K systems have been adopted by Tn7-like transposons to direct RNA-guided ...Canonical CRISPR-Cas systems provide adaptive immunity against mobile genetic elements. However, type I-F, I-B and V-K systems have been adopted by Tn7-like transposons to direct RNA-guided transposon insertion. Type V-K CRISPR-associated transposons rely on the pseudonuclease Cas12k, the transposase TnsB, the AAA+ ATPase TnsC and the zinc-finger protein TniQ, but the molecular mechanism of RNA-directed DNA transposition has remained elusive. Here we report cryo-electron microscopic structures of a Cas12k-guide RNA-target DNA complex and a DNA-bound, polymeric TnsC filament from the CRISPR-associated transposon system of the photosynthetic cyanobacterium Scytonema hofmanni. The Cas12k complex structure reveals an intricate guide RNA architecture and critical interactions mediating RNA-guided target DNA recognition. TnsC helical filament assembly is ATP-dependent and accompanied by structural remodelling of the bound DNA duplex. In vivo transposition assays corroborate key features of the structures, and biochemical experiments show that TniQ restricts TnsC polymerization, while TnsB interacts directly with TnsC filaments to trigger their disassembly upon ATP hydrolysis. Together, these results suggest that RNA-directed target selection by Cas12k primes TnsC polymerization and DNA remodelling, generating a recruitment platform for TnsB to catalyse site-specific transposon insertion. Insights from this work will inform the development of CRISPR-associated transposons as programmable site-specific gene insertion tools. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13489.map.gz emd_13489.map.gz | 152.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13489-v30.xml emd-13489-v30.xml emd-13489.xml emd-13489.xml | 10.7 KB 10.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13489.png emd_13489.png | 76.1 KB | ||
| Filedesc metadata |  emd-13489.cif.gz emd-13489.cif.gz | 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13489 http://ftp.pdbj.org/pub/emdb/structures/EMD-13489 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13489 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13489 | HTTPS FTP |
-Validation report
| Summary document |  emd_13489_validation.pdf.gz emd_13489_validation.pdf.gz | 611.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13489_full_validation.pdf.gz emd_13489_full_validation.pdf.gz | 610.7 KB | Display | |
| Data in XML |  emd_13489_validation.xml.gz emd_13489_validation.xml.gz | 6.9 KB | Display | |
| Data in CIF |  emd_13489_validation.cif.gz emd_13489_validation.cif.gz | 7.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13489 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13489 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13489 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13489 | HTTPS FTP |
-Related structure data
| Related structure data |  7plhMC  7oxdC  7plaC  9go0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13489.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13489.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
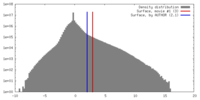
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of ShTnsC with AMPPNP and dsDNA
| Entire | Name: Complex of ShTnsC with AMPPNP and dsDNA |
|---|---|
| Components |
|
-Supramolecule #1: Complex of ShTnsC with AMPPNP and dsDNA
| Supramolecule | Name: Complex of ShTnsC with AMPPNP and dsDNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|
-Macromolecule #1: ShTnsC
| Macromolecule | Name: ShTnsC / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Scytonema hofmannii (bacteria) Scytonema hofmannii (bacteria) |
| Molecular weight | Theoretical: 31.444617 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTEAQAIAKQ LGGVKPDDEW LQAEIARLKG KSIVPLQQVK TLHDWLDGKR KARKSCRVVG ESRTGKTVAC DAYRYRHKPQ QEAGRPPTV PVVYIRPHQK CGPKDLFKKI TEYLKYRVTK GTVSDFRDRT IEVLKGCGVE MLIIDEADRL KPETFADVRD I AEDLGIAV ...String: MTEAQAIAKQ LGGVKPDDEW LQAEIARLKG KSIVPLQQVK TLHDWLDGKR KARKSCRVVG ESRTGKTVAC DAYRYRHKPQ QEAGRPPTV PVVYIRPHQK CGPKDLFKKI TEYLKYRVTK GTVSDFRDRT IEVLKGCGVE MLIIDEADRL KPETFADVRD I AEDLGIAV VLVGTDRLDA VIKRDEQVLE RFRAHLRFGK LSGEDFKNTV EMWEQMVLKL PVSSNLKSKE MLRILTSATE GY IGRLDEI LREAAIRSLS RGLKKIDKAV LQEVAKEYK |
-Macromolecule #2: DNA (5'-D(P*AP*TP*AP*TP*AP*TP*AP*TP*AP*TP*AP*TP*AP*TP*AP*TP*AP*TP...
| Macromolecule | Name: DNA (5'-D(P*AP*TP*AP*TP*AP*TP*AP*TP*AP*TP*AP*TP*AP*TP*AP*TP*AP*TP*AP*TP*AP*T)-3') type: dna / ID: 2 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 6.746438 KDa |
| Sequence | String: (DA)(DT)(DA)(DT)(DA)(DT)(DA)(DT)(DA)(DT) (DA)(DT)(DA)(DT)(DA)(DT)(DA)(DT)(DA)(DT) (DA)(DT) |
-Macromolecule #3: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 3 / Number of copies: 7 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 7 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 66.39 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 6.78 Å Applied symmetry - Helical parameters - Δ&Phi: 59.72 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 3.57 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 77544 |
|---|---|
| Startup model | Type of model: PDB ENTRY |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller











 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)