[English] 日本語
 Yorodumi
Yorodumi- EMDB-13346: Single-particle cryo-EM reconstruction of the tetrahedral 24mer o... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Single-particle cryo-EM reconstruction of the tetrahedral 24mer of Hsp17 from Caenorhabditis elegans | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | small heat shock protein / Caenorhabditis elegans / proteostasis / CHAPERONE | |||||||||
| Function / homology | Alpha crystallin/Small heat shock protein, animal type / Hsp20/alpha crystallin family / Small heat shock protein (sHSP) domain profile. / Alpha crystallin/Hsp20 domain / HSP20-like chaperone / SHSP domain-containing protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
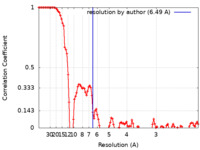
| Method | single particle reconstruction / cryo EM / Resolution: 6.49 Å | |||||||||
 Authors Authors | Rossa B / Weinkauf S | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2023 Journal: J Biol Chem / Year: 2023Title: The permanently chaperone-active small heat shock protein Hsp17 from Caenorhabditis elegans exhibits topological separation of its N-terminal regions. Authors: Annika Strauch / Benjamin Rossa / Fabian Köhler / Simon Haeussler / Moritz Mühlhofer / Florian Rührnößl / Caroline Körösy / Yevheniia Bushman / Barbara Conradt / Martin Haslbeck / ...Authors: Annika Strauch / Benjamin Rossa / Fabian Köhler / Simon Haeussler / Moritz Mühlhofer / Florian Rührnößl / Caroline Körösy / Yevheniia Bushman / Barbara Conradt / Martin Haslbeck / Sevil Weinkauf / Johannes Buchner /   Abstract: Small Heat shock proteins (sHsps) are a family of molecular chaperones that bind nonnative proteins in an ATP-independent manner. Caenorhabditis elegans encodes 16 different sHsps, among them Hsp17, ...Small Heat shock proteins (sHsps) are a family of molecular chaperones that bind nonnative proteins in an ATP-independent manner. Caenorhabditis elegans encodes 16 different sHsps, among them Hsp17, which is evolutionarily distinct from other sHsps in the nematode. The structure and mechanism of Hsp17 and how these may differ from other sHsps remain unclear. Here, we find that Hsp17 has a distinct expression pattern, structural organization, and chaperone function. Consistent with its presence under nonstress conditions, and in contrast to many other sHsps, we determined that Hsp17 is a mono-disperse, permanently active chaperone in vitro, which interacts with hundreds of different C. elegans proteins under physiological conditions. Additionally, our cryo-EM structure of Hsp17 reveals that in the 24-mer complex, 12 N-terminal regions are involved in its chaperone function. These flexible regions are located on the outside of the spherical oligomer, whereas the other 12 N-terminal regions are engaged in stabilizing interactions in its interior. This allows the same region in Hsp17 to perform different functions depending on the topological context. Taken together, our results reveal structural and functional features that further define the structural basis of permanently active sHsps. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13346.map.gz emd_13346.map.gz | 96.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13346-v30.xml emd-13346-v30.xml emd-13346.xml emd-13346.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13346_fsc.xml emd_13346_fsc.xml | 10.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_13346.png emd_13346.png | 82.5 KB | ||
| Masks |  emd_13346_msk_1.map emd_13346_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13346.cif.gz emd-13346.cif.gz | 5.9 KB | ||
| Others |  emd_13346_half_map_1.map.gz emd_13346_half_map_1.map.gz emd_13346_half_map_2.map.gz emd_13346_half_map_2.map.gz | 95.5 MB 95.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13346 http://ftp.pdbj.org/pub/emdb/structures/EMD-13346 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13346 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13346 | HTTPS FTP |
-Related structure data
| Related structure data |  7pe3MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13346.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13346.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_13346_msk_1.map emd_13346_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_13346_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_13346_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tetrahedral oligomer (24mer) of Hsp17
| Entire | Name: Tetrahedral oligomer (24mer) of Hsp17 |
|---|---|
| Components |
|
-Supramolecule #1: Tetrahedral oligomer (24mer) of Hsp17
| Supramolecule | Name: Tetrahedral oligomer (24mer) of Hsp17 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 422 KDa |
-Macromolecule #1: SHSP domain-containing protein
| Macromolecule | Name: SHSP domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.44251 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDRRFPPFSP FFNHGRFFDD VDFDRHMIRP YWADQTMLTG HRVGDAIDVV NNDQEYNVSV DVSQFEPEEL KVNIVDNQLI IEGKHNEKT DKYGQVERHF VRKYNLPTGV RPEQIKSELS NNGVLTVKYE KNQEQQPKSI PITIVPKRN UniProtKB: SHSP domain-containing protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: PBS |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 20 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: waiting time 0, blotting force 5, blotting time 2.5. |
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 2568 / Average electron dose: 55.0 e/Å2 Details: Images were recorded in CDS mode as tif-stacks with 30 frames. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-7pe3: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)