+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13231 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Outer-membrane core complex C1 map | |||||||||
 Map data Map data | Outer Membrane Core Complex C1 map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology | Type IV conjugative transfer system protein TraV / Type IV conjugative transfer system lipoprotein (TraV) / Prokaryotic membrane lipoprotein lipid attachment site profile. / Type IV conjugative transfer system lipoprotein TraV Function and homology information Function and homology information | |||||||||
| Biological species |  Salmonella enterica (bacteria) Salmonella enterica (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.7 Å | |||||||||
 Authors Authors | Amin H / Ilangovan A / Costa TRD | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Architecture of the outer-membrane core complex from a conjugative type IV secretion system. Authors: Himani Amin / Aravindan Ilangovan / Tiago R D Costa /  Abstract: Conjugation is one of the most important processes that bacteria utilize to spread antibiotic resistance genes among bacterial populations. Interbacterial DNA transfer requires a large double ...Conjugation is one of the most important processes that bacteria utilize to spread antibiotic resistance genes among bacterial populations. Interbacterial DNA transfer requires a large double membrane-spanning nanomachine called the type 4 secretion system (T4SS) made up of the inner-membrane complex (IMC), the outer-membrane core complex (OMCC) and the conjugative pilus. The iconic F plasmid-encoded T4SS has been central in understanding conjugation for several decades, however atomic details of its structure are not known. Here, we report the structure of a complete conjugative OMCC encoded by the pED208 plasmid from E. coli, solved by cryo-electron microscopy at 3.3 Å resolution. This 2.1 MDa complex has a unique arrangement with two radial concentric rings, each having a different symmetry eventually contributing to remarkable differences in protein stoichiometry and flexibility in comparison to other OMCCs. Our structure suggests that F-OMCC is a highly dynamic complex, with implications for pilus extension and retraction during conjugation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13231.map.gz emd_13231.map.gz | 398 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13231-v30.xml emd-13231-v30.xml emd-13231.xml emd-13231.xml | 7.8 KB 7.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13231.png emd_13231.png | 40.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13231 http://ftp.pdbj.org/pub/emdb/structures/EMD-13231 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13231 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13231 | HTTPS FTP |
-Validation report
| Summary document |  emd_13231_validation.pdf.gz emd_13231_validation.pdf.gz | 363.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13231_full_validation.pdf.gz emd_13231_full_validation.pdf.gz | 363.2 KB | Display | |
| Data in XML |  emd_13231_validation.xml.gz emd_13231_validation.xml.gz | 7.5 KB | Display | |
| Data in CIF |  emd_13231_validation.cif.gz emd_13231_validation.cif.gz | 8.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13231 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13231 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13231 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13231 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13231.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13231.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Outer Membrane Core Complex C1 map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
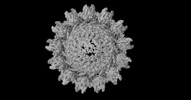
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
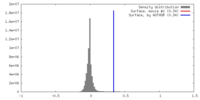
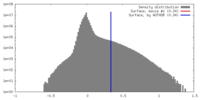
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Outer-membrane core complex C1 map
| Entire | Name: Outer-membrane core complex C1 map |
|---|---|
| Components |
|
-Supramolecule #1: Outer-membrane core complex C1 map
| Supramolecule | Name: Outer-membrane core complex C1 map / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Salmonella enterica (bacteria) Salmonella enterica (bacteria) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: Ab initio |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 5.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 95177 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)