[English] 日本語
 Yorodumi
Yorodumi- EMDB-13164: Cryo-EM structure of encapsulin from Mycobacterium tuberculosis -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of encapsulin from Mycobacterium tuberculosis | |||||||||
 Map data Map data | LocSpiral map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | encapsulin / cfp29 / nanocompartment / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | Type 1 encapsulin shell protein / Encapsulating protein for peroxidase / : / encapsulin nanocompartment / hydrolase activity / plasma membrane / Type 1 encapsulin shell protein Function and homology information Function and homology information | |||||||||
| Biological species |  Mycobacterium tuberculosis H37Rv (bacteria) / Mycobacterium tuberculosis H37Rv (bacteria) /  | |||||||||
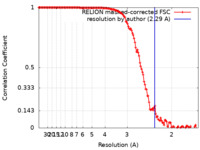
| Method | single particle reconstruction / cryo EM / Resolution: 2.29 Å | |||||||||
 Authors Authors | Lewis CJ / Berger C | |||||||||
| Funding support |  Netherlands, 2 items Netherlands, 2 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2025 Journal: Commun Biol / Year: 2025Title: In situ and in vitro cryo-EM reveal structures of mycobacterial encapsulin assembly intermediates. Authors: Casper Berger / Chris Lewis / Ye Gao / Kèvin Knoops / Carmen López-Iglesias / Peter J Peters / Raimond B G Ravelli /   Abstract: Prokaryotes rely on proteinaceous compartments such as encapsulin to isolate harmful reactions. Encapsulin are widely expressed by bacteria, including the Mycobacteriaceae, which include the human ...Prokaryotes rely on proteinaceous compartments such as encapsulin to isolate harmful reactions. Encapsulin are widely expressed by bacteria, including the Mycobacteriaceae, which include the human pathogens Mycobacterium tuberculosis and Mycobacterium leprae. Structures of fully assembled encapsulin shells have been determined for several species, but encapsulin assembly and cargo encapsulation are still poorly characterised, because of the absence of encapsulin structures in intermediate assembly states. We combine in situ and in vitro structural electron microscopy to show that encapsulins are dynamic assemblies with intermediate states of cargo encapsulation and shell assembly. Using cryo-focused ion beam (FIB) lamella preparation and cryo-electron tomography (CET), we directly visualise encapsulins in Mycobacterium marinum, and observed ribbon-like attachments to the shell, encapsulin shells with and without cargoes, and encapsulin shells in partially assembled states. In vitro cryo-electron microscopy (EM) single-particle analysis of the Mycobacterium tuberculosis encapsulin was used to obtain three structures of the encapsulin shell in intermediate states, as well as a 2.3 Å structure of the fully assembled shell. Based on the analysis of the intermediate encapsulin shell structures, we propose a model of encapsulin self-assembly via the pairwise addition of monomers. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13164.map.gz emd_13164.map.gz | 41.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13164-v30.xml emd-13164-v30.xml emd-13164.xml emd-13164.xml | 23 KB 23 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13164_fsc.xml emd_13164_fsc.xml | 13.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_13164.png emd_13164.png | 237.9 KB | ||
| Masks |  emd_13164_msk_1.map emd_13164_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13164.cif.gz emd-13164.cif.gz | 6.9 KB | ||
| Others |  emd_13164_additional_1.map.gz emd_13164_additional_1.map.gz emd_13164_half_map_1.map.gz emd_13164_half_map_1.map.gz emd_13164_half_map_2.map.gz emd_13164_half_map_2.map.gz | 40.4 MB 170.7 MB 170.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13164 http://ftp.pdbj.org/pub/emdb/structures/EMD-13164 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13164 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13164 | HTTPS FTP |
-Related structure data
| Related structure data |  7p1tMC  9gotC  9hq7C  9hqcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13164.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13164.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LocSpiral map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.834 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_13164_msk_1.map emd_13164_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: relion postprocess masked
| File | emd_13164_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | relion postprocess masked | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halfmap
| File | emd_13164_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: halfmap
| File | emd_13164_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | halfmap | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : encapsulin shell from Mycobacterium tuberculosis
| Entire | Name: encapsulin shell from Mycobacterium tuberculosis |
|---|---|
| Components |
|
-Supramolecule #1: encapsulin shell from Mycobacterium tuberculosis
| Supramolecule | Name: encapsulin shell from Mycobacterium tuberculosis / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
| Molecular weight | Theoretical: 1.729 MDa |
-Macromolecule #1: 29 kDa antigen, Cfp29
| Macromolecule | Name: 29 kDa antigen, Cfp29 / type: protein_or_peptide / ID: 1 / Number of copies: 60 / Enantiomer: LEVO / EC number: Hydrolases; Acting on peptide bonds (peptidases) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.863346 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNNLYRDLAP VTEAAWAEIE LEAARTFKRH IAGRRVVDVS DPGGPVTAAV STGRLIDVKA PTNGVIAHLR ASKPLVRLRV PFTLSRNEI DDVERGSKDS DWEPVKEAAK KLAFVEDRTI FEGYSAASIE GIRSASSNPA LTLPEDPREI PDVISQALSE L RLAGVDGP ...String: MNNLYRDLAP VTEAAWAEIE LEAARTFKRH IAGRRVVDVS DPGGPVTAAV STGRLIDVKA PTNGVIAHLR ASKPLVRLRV PFTLSRNEI DDVERGSKDS DWEPVKEAAK KLAFVEDRTI FEGYSAASIE GIRSASSNPA LTLPEDPREI PDVISQALSE L RLAGVDGP YSVLLSADVY TKVSETSDHG YPIREHLNRL VDGDIIWAPA IDGAFVLTTR GGDFDLQLGT DVAIGYASHD TD TVRLYLQ ETLTFLCYTA EASVALSH UniProtKB: Type 1 encapsulin shell protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average exposure time: 1.8 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 163000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)