[English] 日本語
 Yorodumi
Yorodumi- EMDB-13136: Subtomogram average of authentic mumps virus nucleocapsid from He... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram average of authentic mumps virus nucleocapsid from HeLa cell lysate of short helical pitch | |||||||||
 Map data Map data | Minor conformation of authentic mumps virus nucleocapsid from the lysate of HeLa cells after stress treatment by subtomogram averaging | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Helical filament / Nucleocapsid / Protein-RNA complex / Scaffold / VIRUS | |||||||||
| Biological species |  Mumps virus genotype A Mumps virus genotype A | |||||||||
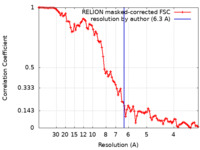
| Method | subtomogram averaging / cryo EM / Resolution: 6.3 Å | |||||||||
 Authors Authors | Mahamid J / Zhang X / Pflaesterer T | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Molecular mechanisms of stress-induced reactivation in mumps virus condensates. Authors: Xiaojie Zhang / Sindhuja Sridharan / Ievgeniia Zagoriy / Christina Eugster Oegema / Cyan Ching / Tim Pflaesterer / Herman K H Fung / Isabelle Becher / Ina Poser / Christoph W Müller / ...Authors: Xiaojie Zhang / Sindhuja Sridharan / Ievgeniia Zagoriy / Christina Eugster Oegema / Cyan Ching / Tim Pflaesterer / Herman K H Fung / Isabelle Becher / Ina Poser / Christoph W Müller / Anthony A Hyman / Mikhail M Savitski / Julia Mahamid /  Abstract: Negative-stranded RNA viruses can establish long-term persistent infection in the form of large intracellular inclusions in the human host and cause chronic diseases. Here, we uncover how cellular ...Negative-stranded RNA viruses can establish long-term persistent infection in the form of large intracellular inclusions in the human host and cause chronic diseases. Here, we uncover how cellular stress disrupts the metastable host-virus equilibrium in persistent infection and induces viral replication in a culture model of mumps virus. Using a combination of cell biology, whole-cell proteomics, and cryo-electron tomography, we show that persistent viral replication factories are dynamic condensates and identify the largely disordered viral phosphoprotein as a driver of their assembly. Upon stress, increased phosphorylation of the phosphoprotein at its interaction interface with the viral polymerase coincides with the formation of a stable replication complex. By obtaining atomic models for the authentic mumps virus nucleocapsid, we elucidate a concomitant conformational change that exposes the viral genome to its replication machinery. These events constitute a stress-mediated switch within viral condensates that provide an environment to support upregulation of viral replication. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13136.map.gz emd_13136.map.gz | 8.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13136-v30.xml emd-13136-v30.xml emd-13136.xml emd-13136.xml | 12.6 KB 12.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13136_fsc.xml emd_13136_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_13136.png emd_13136.png | 40.3 KB | ||
| Filedesc metadata |  emd-13136.cif.gz emd-13136.cif.gz | 5.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13136 http://ftp.pdbj.org/pub/emdb/structures/EMD-13136 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13136 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13136 | HTTPS FTP |
-Validation report
| Summary document |  emd_13136_validation.pdf.gz emd_13136_validation.pdf.gz | 460.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13136_full_validation.pdf.gz emd_13136_full_validation.pdf.gz | 460 KB | Display | |
| Data in XML |  emd_13136_validation.xml.gz emd_13136_validation.xml.gz | 11.1 KB | Display | |
| Data in CIF |  emd_13136_validation.cif.gz emd_13136_validation.cif.gz | 14.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13136 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13136 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13136 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13136 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13136.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13136.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Minor conformation of authentic mumps virus nucleocapsid from the lysate of HeLa cells after stress treatment by subtomogram averaging | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.6938 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Authentic Mumps virus nucleocapsid-RNA complex
| Entire | Name: Authentic Mumps virus nucleocapsid-RNA complex |
|---|---|
| Components |
|
-Supramolecule #1: Authentic Mumps virus nucleocapsid-RNA complex
| Supramolecule | Name: Authentic Mumps virus nucleocapsid-RNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mumps virus genotype A Mumps virus genotype A |
-Macromolecule #1: Mumps virus nucleocapsid
| Macromolecule | Name: Mumps virus nucleocapsid / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mumps virus genotype A / Strain: Enders Mumps virus genotype A / Strain: Enders |
| Sequence | String: MSSVLKAFER FTIEQELQDR GEEGSIPPET LKSAVKVFVI NTPNPTTRYQ MLNFCLRIIC SQNARASHRV GALITLFSLP SAGMQNHIRL ADRSPEAQIE RCEIDGFEPG TYRLIPNARA NLTANEIAAY ALLADDLPPT INNGTPYVHA DVEGQPCDEI EQFLDRCYSV ...String: MSSVLKAFER FTIEQELQDR GEEGSIPPET LKSAVKVFVI NTPNPTTRYQ MLNFCLRIIC SQNARASHRV GALITLFSLP SAGMQNHIRL ADRSPEAQIE RCEIDGFEPG TYRLIPNARA NLTANEIAAY ALLADDLPPT INNGTPYVHA DVEGQPCDEI EQFLDRCYSV LIQAWVMVCK CMTAYDQPAG SADRRFAKYQ QQGRLEARYM LQPEAQRLIQ TAIRKSLVVR QYLTFELQLA RRQGLLSNRY YAMVGDIGKY IENSGLTAFF LTLKYALGTK WSPLSLAAFT GELTKLRSLM MLYRDIGEQA RYLALLEAPQ IMDFAPGGYP LIFSYAMGVG TVLDAQMRNY TYARPFLNGY YFQIGVETAR RQQGTVDNRV ADDLGLTPEQ RTEVTQLVDR LARGRGAGIP GGPVNPFVPP VQQQQPAAVY ADIPALEESD DDGDEDGGAG FQNGVQVPAV RQGGQTDFRA QPLQDPIQAQ LFMPLYPQVS NIPSNQNHQI NRIGGLENQD LLRYNENGDS QQDARGEHGN TFPNNPNQNA QLQVGDWDE |
-Macromolecule #2: RNA
| Macromolecule | Name: RNA / type: rna / ID: 2 |
|---|---|
| Source (natural) | Organism:  Mumps virus genotype A Mumps virus genotype A |
| Sequence | String: UUUUUU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Average electron dose: 2.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)