[English] 日本語
 Yorodumi
Yorodumi- EMDB-13008: Mechanosensitive channel MscS solubilized with LMNG in open confo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13008 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mechanosensitive channel MscS solubilized with LMNG in open conformation with added lipid | |||||||||
 Map data Map data | post-processed: unmasked, sharpened | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mechanosensitive channel / LMNG / delipidation / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
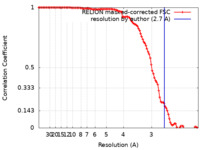
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Rasmussen T / Flegler VJ | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Mechanosensitive channel gating by delipidation. Authors: Vanessa Judith Flegler / Akiko Rasmussen / Karina Borbil / Lea Boten / Hsuan-Ai Chen / Hanna Deinlein / Julia Halang / Kristin Hellmanzik / Jessica Löffler / Vanessa Schmidt / Cihan Makbul ...Authors: Vanessa Judith Flegler / Akiko Rasmussen / Karina Borbil / Lea Boten / Hsuan-Ai Chen / Hanna Deinlein / Julia Halang / Kristin Hellmanzik / Jessica Löffler / Vanessa Schmidt / Cihan Makbul / Christian Kraft / Rainer Hedrich / Tim Rasmussen / Bettina Böttcher /  Abstract: The mechanosensitive channel of small conductance (MscS) protects bacteria against hypoosmotic shock. It can sense the tension in the surrounding membrane and releases solutes if the pressure in the ...The mechanosensitive channel of small conductance (MscS) protects bacteria against hypoosmotic shock. It can sense the tension in the surrounding membrane and releases solutes if the pressure in the cell is getting too high. The membrane contacts MscS at sensor paddles, but lipids also leave the membrane and move along grooves between the paddles to reside as far as 15 Å away from the membrane in hydrophobic pockets. One sensing model suggests that a higher tension pulls lipids from the grooves back to the membrane, which triggers gating. However, it is still unclear to what degree this model accounts for sensing and what contribution the direct interaction of the membrane with the channel has. Here, we show that MscS opens when it is sufficiently delipidated by incubation with the detergent dodecyl-β-maltoside or the branched detergent lauryl maltose neopentyl glycol. After addition of detergent-solubilized lipids, it closes again. These results support the model that lipid extrusion causes gating: Lipids are slowly removed from the grooves and pockets by the incubation with detergent, which triggers opening. Addition of lipids in micelles allows lipids to migrate back into the pockets, which closes the channel even in the absence of a membrane. Based on the distribution of the aliphatic chains in the open and closed conformation, we propose that during gating, lipids leave the complex on the cytosolic leaflet at the height of highest lateral tension, while on the periplasmic side, lipids flow into gaps, which open between transmembrane helices. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13008.map.gz emd_13008.map.gz | 96.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13008-v30.xml emd-13008-v30.xml emd-13008.xml emd-13008.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13008_fsc.xml emd_13008_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_13008.png emd_13008.png | 184.3 KB | ||
| Masks |  emd_13008_msk_1.map emd_13008_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13008.cif.gz emd-13008.cif.gz | 6.5 KB | ||
| Others |  emd_13008_half_map_1.map.gz emd_13008_half_map_1.map.gz emd_13008_half_map_2.map.gz emd_13008_half_map_2.map.gz | 81.1 MB 81.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13008 http://ftp.pdbj.org/pub/emdb/structures/EMD-13008 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13008 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13008 | HTTPS FTP |
-Validation report
| Summary document |  emd_13008_validation.pdf.gz emd_13008_validation.pdf.gz | 877.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13008_full_validation.pdf.gz emd_13008_full_validation.pdf.gz | 876.7 KB | Display | |
| Data in XML |  emd_13008_validation.xml.gz emd_13008_validation.xml.gz | 18.1 KB | Display | |
| Data in CIF |  emd_13008_validation.cif.gz emd_13008_validation.cif.gz | 23.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13008 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13008 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13008 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13008 | HTTPS FTP |
-Related structure data
| Related structure data |  7ooaMC  7onjC  7onlC  7oo0C  7oo6C  7oo8C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13008.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13008.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | post-processed: unmasked, sharpened | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0635 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
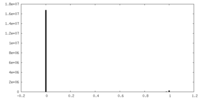
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13008_msk_1.map emd_13008_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
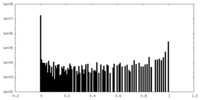
| Density Histograms |
-Half map: unfiltered half-map
| File | emd_13008_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered half-map | ||||||||||||
| Projections & Slices |
| ||||||||||||
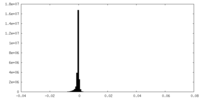
| Density Histograms |
-Half map: unfiltered half-map
| File | emd_13008_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered half-map | ||||||||||||
| Projections & Slices |
| ||||||||||||
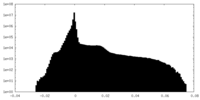
| Density Histograms |
- Sample components
Sample components
-Entire : homoheptameric complex of MscS coordinated with lipids and deterg...
| Entire | Name: homoheptameric complex of MscS coordinated with lipids and detergents DDM and LMNG |
|---|---|
| Components |
|
-Supramolecule #1: homoheptameric complex of MscS coordinated with lipids and deterg...
| Supramolecule | Name: homoheptameric complex of MscS coordinated with lipids and detergents DDM and LMNG type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 224 KDa |
-Macromolecule #1: Mechanosensitive channel of small conductance (MscS)
| Macromolecule | Name: Mechanosensitive channel of small conductance (MscS) / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: Escherichia coli SE11 |
| Molecular weight | Theoretical: 31.994039 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEDLNVVDSI NGAGSWLVAN QALLLSYAVN IVAALAIIIV GLIIARMISN AVNRLMISRK IDATVADFLS ALVRYGIIAF TLIAALGRV GVQTASVIAV LGAAGLAVGL ALQGSLSNLA AGVLLVMFRP FRAGEYVDLG GVAGTVLSVQ IFSTTMRTAD G KIIVIPNG ...String: MEDLNVVDSI NGAGSWLVAN QALLLSYAVN IVAALAIIIV GLIIARMISN AVNRLMISRK IDATVADFLS ALVRYGIIAF TLIAALGRV GVQTASVIAV LGAAGLAVGL ALQGSLSNLA AGVLLVMFRP FRAGEYVDLG GVAGTVLSVQ IFSTTMRTAD G KIIVIPNG KIIAGNIINF SREPVRRNEF IIGVAYDSDI DQVKQILTNI IQSEDRILKD REMTVRLNEL GASSINFVVR VW SNSGDLQ NVYWDVLERI KREFDAAGIS FPYPQMDVNF KRVKEDKAAL EHHHHHH UniProtKB: Uncharacterized protein |
-Macromolecule #2: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
| Macromolecule | Name: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine / type: ligand / ID: 2 / Number of copies: 7 / Formula: PEE |
|---|---|
| Molecular weight | Theoretical: 744.034 Da |
| Chemical component information |  ChemComp-PEE: |
-Macromolecule #3: Lauryl Maltose Neopentyl Glycol
| Macromolecule | Name: Lauryl Maltose Neopentyl Glycol / type: ligand / ID: 3 / Number of copies: 21 / Formula: AV0 |
|---|---|
| Molecular weight | Theoretical: 1.005188 KDa |
| Chemical component information |  ChemComp-AV0: |
-Macromolecule #4: DODECYL-BETA-D-MALTOSIDE
| Macromolecule | Name: DODECYL-BETA-D-MALTOSIDE / type: ligand / ID: 4 / Number of copies: 7 / Formula: LMT |
|---|---|
| Molecular weight | Theoretical: 510.615 Da |
| Chemical component information |  ChemComp-LMT: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 150 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 7 sec blotting, blot force +25. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 10080 / Average exposure time: 75.0 sec. / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: WEIGHTED MAP SUM AT ATOM CENTERS |
| Output model |  PDB-7ooa: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)