+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12986 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Control sample rancRNA_18 | ||||||||||||
 Map data Map data | postrpocessed 60S all particles | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.1 Å | ||||||||||||
 Authors Authors | Zuber B / Iacovache I | ||||||||||||
| Funding support |  Switzerland, 3 items Switzerland, 3 items
| ||||||||||||
 Citation Citation |  Journal: RNA Biol / Year: 2021 Journal: RNA Biol / Year: 2021Title: A small ribosome-associated ncRNA globally inhibits translation by restricting ribosome dynamics. Authors: Julia Reuther / Lukas Schneider / Ioan Iacovache / Andreas Pircher / Walid H Gharib / Benoît Zuber / Norbert Polacek /  Abstract: Ribosome-associated non-coding RNAs (rancRNAs) have been recognized as an emerging class of regulatory molecules capable of fine-tuning translation in all domains of life. RancRNAs are ideally suited ...Ribosome-associated non-coding RNAs (rancRNAs) have been recognized as an emerging class of regulatory molecules capable of fine-tuning translation in all domains of life. RancRNAs are ideally suited for allowing a swift response to changing environments and are therefore considered pivotal during the first wave of stress adaptation. Previously, we identified an mRNA-derived 18 nucleotides long rancRNA (rancRNA_18) in that rapidly downregulates protein synthesis during hyperosmotic stress. However, the molecular mechanism of action remained enigmatic. Here, we combine biochemical, genetic, transcriptome-wide and structural evidence, thus revealing rancRNA_18 as global translation inhibitor by targeting the E-site region of the large ribosomal subunit. Ribosomes carrying rancRNA_18 possess decreased affinity for A-site tRNA and impaired structural dynamics. Cumulatively, these discoveries reveal the mode of action of a rancRNA involved in modulating protein biosynthesis at a thus far unequalled precision. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
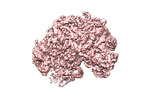
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12986.map.gz emd_12986.map.gz | 25.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12986-v30.xml emd-12986-v30.xml emd-12986.xml emd-12986.xml | 21.4 KB 21.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12986_fsc.xml emd_12986_fsc.xml | 16.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_12986.png emd_12986.png | 82.4 KB | ||
| Others |  emd_12986_additional_1.map.gz emd_12986_additional_1.map.gz emd_12986_additional_2.map.gz emd_12986_additional_2.map.gz emd_12986_additional_3.map.gz emd_12986_additional_3.map.gz emd_12986_additional_4.map.gz emd_12986_additional_4.map.gz emd_12986_additional_5.map.gz emd_12986_additional_5.map.gz | 274.1 MB 275.6 MB 274.8 MB 275 MB 273.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12986 http://ftp.pdbj.org/pub/emdb/structures/EMD-12986 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12986 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12986 | HTTPS FTP |
-Validation report
| Summary document |  emd_12986_validation.pdf.gz emd_12986_validation.pdf.gz | 344.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12986_full_validation.pdf.gz emd_12986_full_validation.pdf.gz | 344.4 KB | Display | |
| Data in XML |  emd_12986_validation.xml.gz emd_12986_validation.xml.gz | 15.1 KB | Display | |
| Data in CIF |  emd_12986_validation.cif.gz emd_12986_validation.cif.gz | 20.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12986 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12986 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12986 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12986 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12986.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12986.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | postrpocessed 60S all particles | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.052 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
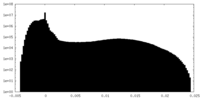
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: 2D classification C5
| File | emd_12986_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 2D classification C5 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 2D classification C6
| File | emd_12986_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 2D classification C6 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Refinement 80s all particles
| File | emd_12986_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement 80s all particles | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 2D classification C10
| File | emd_12986_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 2D classification C10 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 2D classification C8
| File | emd_12986_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 2D classification C8 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : control sample for rancRNA_18 - cryoEM analysis and classification
| Entire | Name: control sample for rancRNA_18 - cryoEM analysis and classification |
|---|---|
| Components |
|
-Supramolecule #1: control sample for rancRNA_18 - cryoEM analysis and classification
| Supramolecule | Name: control sample for rancRNA_18 - cryoEM analysis and classification type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.6 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 0.2 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.25 mm / Nominal magnification: 100000 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)