[English] 日本語
 Yorodumi
Yorodumi- EMDB-1294: Three-dimensional structure of the native spliceosome by cryo-ele... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1294 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Three-dimensional structure of the native spliceosome by cryo-electron microscopy. | |||||||||
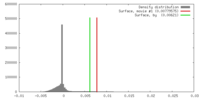
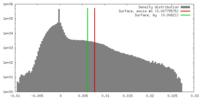
 Map data Map data | Biological isosurface is at density value 0.007 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 22.0 Å | |||||||||
 Authors Authors | Azubel M / Wolf SG / Sperling J / Sperling R | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2004 Journal: Mol Cell / Year: 2004Title: Three-dimensional structure of the native spliceosome by cryo-electron microscopy. Authors: Maia Azubel / Sharon G Wolf / Joseph Sperling / Ruth Sperling /  Abstract: Splicing of pre-mRNA occurs in a multicomponent macromolecular machine--the spliceosome. The spliceosome can be assembled in vitro by a stepwise assembly of a number of snRNPs and additional proteins ...Splicing of pre-mRNA occurs in a multicomponent macromolecular machine--the spliceosome. The spliceosome can be assembled in vitro by a stepwise assembly of a number of snRNPs and additional proteins on exogenously added pre-mRNA. In contrast, splicing in vivo occurs in preformed particles where endogenous pre-mRNAs are packaged with all five spliceosomal U snRNPs (penta-snRNP) together with other splicing factors. Here we present a three-dimensional image reconstruction by cryo-electron microscopy of native spliceosomes, derived from cell nuclei, at a resolution of 20 angstroms. The structure revealed an elongated globular particle made up of two distinct subunits connected to each other leaving a tunnel in between. We show here that the larger subunit is a suitable candidate to accommodate the penta-snRNP, and that the tunnel could accommodate the pre-mRNA component of the spliceosome. The features this structure reveals provide new insight into the global architecture of the native splicing machine. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1294.map.gz emd_1294.map.gz | 3.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1294-v30.xml emd-1294-v30.xml emd-1294.xml emd-1294.xml | 9 KB 9 KB | Display Display |  EMDB header EMDB header |
| Images |  1294.gif 1294.gif | 31.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1294 http://ftp.pdbj.org/pub/emdb/structures/EMD-1294 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1294 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1294 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1294.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1294.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Biological isosurface is at density value 0.007 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.25 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : native spliceosome
| Entire | Name: native spliceosome |
|---|---|
| Components |
|
-Supramolecule #1000: native spliceosome
| Supramolecule | Name: native spliceosome / type: sample / ID: 1000 / Oligomeric state: monomeric / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 4.8 MDa / Method: STEM mass measurement |
-Supramolecule #1: native spliceosome
| Supramolecule | Name: native spliceosome / type: organelle_or_cellular_component / ID: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: human / Cell: HeLa / Organelle: Nucleus / Location in cell: nucleus Homo sapiens (human) / synonym: human / Cell: HeLa / Organelle: Nucleus / Location in cell: nucleus |
| Molecular weight | Theoretical: 4.8 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Grid | Details: lacey (SPI) or Quantifoil grids |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: I. Talmon plunger Method: the samples were incubated in Teflon wells under charged lipid monolayers for 20 min., picked up on grids, rinsed, blotted and plunged into liquid ethane. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 95 K / Max: 100 K / Average: 97 K |
| Alignment procedure | Legacy - Astigmatism: correction at 100,000 to 250,000 times mag |
| Details | magnification with calibrated camera postmag factor was 93,620 |
| Image recording | Category: CCD / Film or detector model: GENERIC TVIPS / Digitization - Sampling interval: 24 µm / Average electron dose: 10 e/Å2 / Details: TVIPS Biocam 1k X 1k CCD camera / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 62000 |
| Sample stage | Specimen holder: side entry liquid nitrogen-cooled cryo specimen holder Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: phase correction, each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 22.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Spider Details: see Supplemental material at Molecular Cell website Number images used: 7500 |
| Final angle assignment | Details: see Supplemental material at Molecular Cell website. |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)