+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Enterococcus faecalis EfrCD in complex with a nanobody | ||||||||||||
 Map data Map data | EfrCD volume | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ABC transporter / Nanobody / Enterococcus faecalis / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationoligopeptide export from mitochondrion / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances / ABC-type oligopeptide transporter activity / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |   | ||||||||||||
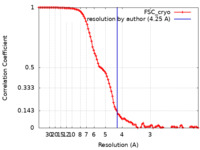
| Method | single particle reconstruction / cryo EM / Resolution: 4.25 Å | ||||||||||||
 Authors Authors | Ehrenbolger K / Hutter CAJ / Meier G / Seeger MA / Barandun J | ||||||||||||
| Funding support |  Sweden, Sweden,  Switzerland, 3 items Switzerland, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2023 Journal: Nat Chem Biol / Year: 2023Title: Deep mutational scan of a drug efflux pump reveals its structure-function landscape. Authors: Gianmarco Meier / Sujani Thavarasah / Kai Ehrenbolger / Cedric A J Hutter / Lea M Hürlimann / Jonas Barandun / Markus A Seeger /   Abstract: Drug efflux is a common resistance mechanism found in bacteria and cancer cells, but studies providing comprehensive functional insights are scarce. In this study, we performed deep mutational ...Drug efflux is a common resistance mechanism found in bacteria and cancer cells, but studies providing comprehensive functional insights are scarce. In this study, we performed deep mutational scanning (DMS) on the bacterial ABC transporter EfrCD to determine the drug efflux activity profile of more than 1,430 single variants. These systematic measurements revealed that the introduction of negative charges at different locations within the large substrate binding pocket results in strongly increased efflux activity toward positively charged ethidium, whereas additional aromatic residues did not display the same effect. Data analysis in the context of an inward-facing cryogenic electron microscopy structure of EfrCD uncovered a high-affinity binding site, which releases bound drugs through a peristaltic transport mechanism as the transporter transits to its outward-facing conformation. Finally, we identified substitutions resulting in rapid Hoechst influx without affecting the efflux activity for ethidium and daunorubicin. Hence, single mutations can convert EfrCD into a drug-specific ABC importer. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12816.map.gz emd_12816.map.gz | 93.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12816-v30.xml emd-12816-v30.xml emd-12816.xml emd-12816.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12816_fsc.xml emd_12816_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_12816.png emd_12816.png | 98.7 KB | ||
| Filedesc metadata |  emd-12816.cif.gz emd-12816.cif.gz | 7.2 KB | ||
| Others |  emd_12816_half_map_1.map.gz emd_12816_half_map_1.map.gz emd_12816_half_map_2.map.gz emd_12816_half_map_2.map.gz | 91.9 MB 91.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12816 http://ftp.pdbj.org/pub/emdb/structures/EMD-12816 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12816 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12816 | HTTPS FTP |
-Related structure data
| Related structure data |  7ocyMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12816.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12816.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EfrCD volume | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: EfrCD half map2
| File | emd_12816_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EfrCD half map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: EfrCD half map1
| File | emd_12816_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EfrCD half map1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : EfrCD in complex with a nanobody
| Entire | Name: EfrCD in complex with a nanobody |
|---|---|
| Components |
|
-Supramolecule #1: EfrCD in complex with a nanobody
| Supramolecule | Name: EfrCD in complex with a nanobody / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Enterococcus faecalis
| Supramolecule | Name: Enterococcus faecalis / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: nanobody
| Supramolecule | Name: nanobody / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ABC transporter ATP-binding protein
| Macromolecule | Name: ABC transporter ATP-binding protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 62.905289 KDa |
| Recombinant expression | Organism:  Lactococcus lactis (lactic acid bacteria) Lactococcus lactis (lactic acid bacteria) |
| Sequence | String: MDLIIQHAKK YKGSVVIALL AVIVMVVSAL WQPKLLQQVL EAIMNDDSDK MKNLGIQLIA IAGLGLVAGV INTIFSAKVA QGVSADIRE ATFRKIQTFS FGNIEKFSAG NLVVRLTNDV TQIQNVIMIA LQTLFRIPFL FIGSFILAML TLPQLWWVIV A LVIAVILI ...String: MDLIIQHAKK YKGSVVIALL AVIVMVVSAL WQPKLLQQVL EAIMNDDSDK MKNLGIQLIA IAGLGLVAGV INTIFSAKVA QGVSADIRE ATFRKIQTFS FGNIEKFSAG NLVVRLTNDV TQIQNVIMIA LQTLFRIPFL FIGSFILAML TLPQLWWVIV A LVIAVILI SMLSFSQMGK HFMIIQNLID KINGIAKENL LGIRVVKSFV QEKNQLSRFT KVSEELTTHN LIVGSLFAVM IP AFMLVAN LAVVGSIFFV SNLVKDDPTL IGGVASFMNY LMQIMMAIII GGMMMMMTSR AAVSIKRIKE VMETEPDVTY KKV PEQELI GSVEFDHVSF RYPGDEEDTL KDISFSIQPG EMIGIVGATG AGKSTLAQLI PRLFDPTEGK IEVGGVDLRE VNEH SLRKT VSFVLQKAIL FSGTIAQNLR HGKRDASEAD MERASGIAQA KEFIEKLAEG YDAPVEERSN NFSGGQKQRL SITRG VIGE PKILILDDST SALDARSERL VREALDKELK ETTTIVIAQK ISSVVHADRI LVLDNGRLVG EGTHEELAAT NPVYQE IYE TQKGKEEA UniProtKB: ABC transporter ATP-binding protein |
-Macromolecule #2: ABC transporter ATP-binding protein
| Macromolecule | Name: ABC transporter ATP-binding protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 66.213734 KDa |
| Recombinant expression | Organism:  Lactococcus lactis (lactic acid bacteria) Lactococcus lactis (lactic acid bacteria) |
| Sequence | String: MTDLIKASKF FYHYLKRYKV SFLFIFLAIF AATYLQVKAP QFVGEAIQEL AKYAVNVMQG KDDKSAFVSV IWKLLIFYVL TSAASFIYS ILFTQVVGKS TNRMRIGLFN KLEKLTIRFF DSHQDGEILS RFTSDLDNIQ NSLNQALLQV LTNIALLVGV L IMMFRQNV ...String: MTDLIKASKF FYHYLKRYKV SFLFIFLAIF AATYLQVKAP QFVGEAIQEL AKYAVNVMQG KDDKSAFVSV IWKLLIFYVL TSAASFIYS ILFTQVVGKS TNRMRIGLFN KLEKLTIRFF DSHQDGEILS RFTSDLDNIQ NSLNQALLQV LTNIALLVGV L IMMFRQNV ELAWATIAST PIAILIAVFV ISKARKYVDL QQDEVGKLNG YMDEKISGQR VIITNGLQEE TIDGFLEQNE KV RAATYKG QVYSGLLFPM MQGMSLVNTA IVIFFGGWLA INGSVDRAAA LGLVVMFVQY SQQYYQPLMQ ISSGYSMIQL AVT GARRLN EMFDEPDEIR PENGEKLEEI NKAVALNHVV FGYNPETPVL KDVSIHVDKG EMVALVGPTG SGKTTIMNLM NRFY DVNEG AVTFDGVDIR EMDLDSLRSH VGIVLQESVL FSGTIRENIA FGKPEATDEE IVQAAKQANI HEFIVNLEQG YDTEI TEEN NLFSTGQKQL VSIARTIITN PELLILDEAT SNVDTVTEAK IQKAMDEAIK GRTSFVIAHR LKTILNADRI IVLRDG EVI EEGNHHELVE QDGFYAELYK NQFVFE UniProtKB: ABC transporter ATP-binding protein |
-Macromolecule #3: Nanobody
| Macromolecule | Name: Nanobody / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.718138 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPSQLQLVES GGGLVQAGDT LRLSCEASRS FNRMGWYRQA PGKQRDMVAH IFSDGRTRYA DSVQGRFTIS RDNAKNTVYL QMNNLKPED TAVYYCNGFF IQDFWGQGTP VTVSA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV Details: blot force of -5, waiting time of 1 second, blot time of 4.5 seconds at 4C. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Number real images: 3135 / Average electron dose: 52.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.3000000000000003 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)