+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12577 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ColicinE9 intact translocation complex | |||||||||
 Map data Map data | Output map from cryosparc local refinement job, sharpened using deepEMhancer tight target model | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacteriocin complex / import / membrane / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationABC-type vitamin B12 transporter activity / cell cycle / extrachromosomal circular DNA / colicin transmembrane transporter activity / cobalamin transport / protein import / transmembrane transporter complex / porin activity / pore complex / protein homotrimerization ...ABC-type vitamin B12 transporter activity / cell cycle / extrachromosomal circular DNA / colicin transmembrane transporter activity / cobalamin transport / protein import / transmembrane transporter complex / porin activity / pore complex / protein homotrimerization / monoatomic ion channel complex / monoatomic ion channel activity / cell outer membrane / lipopolysaccharide binding / disordered domain specific binding / protein transport / monoatomic ion transmembrane transport / endonuclease activity / killing of cells of another organism / Hydrolases; Acting on ester bonds / periplasmic space / defense response to bacterium / protein domain specific binding / cell division / calcium ion binding / lipid binding / protein-containing complex / metal ion binding / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |    | |||||||||
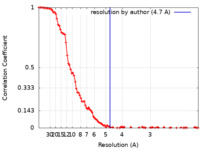
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Webby MN / Kleanthous C | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2021 Journal: EMBO J / Year: 2021Title: Porin threading drives receptor disengagement and establishes active colicin transport through Escherichia coli OmpF. Authors: Marie-Louise R Francis / Melissa N Webby / Nicholas G Housden / Renata Kaminska / Emma Elliston / Boonyaporn Chinthammit / Natalya Lukoyanova / Colin Kleanthous /  Abstract: Bacteria deploy weapons to kill their neighbours during competition for resources and to aid survival within microbiomes. Colicins were the first such antibacterial system identified, yet how these ...Bacteria deploy weapons to kill their neighbours during competition for resources and to aid survival within microbiomes. Colicins were the first such antibacterial system identified, yet how these bacteriocins cross the outer membrane (OM) of Escherichia coli is unknown. Here, by solving the structures of translocation intermediates via cryo-EM and by imaging toxin import, we uncover the mechanism by which the Tol-dependent nuclease colicin E9 (ColE9) crosses the bacterial OM. We show that threading of ColE9's disordered N-terminal domain through two pores of the trimeric porin OmpF causes the colicin to disengage from its primary receptor, BtuB, and reorganises the translocon either side of the membrane. Subsequent import of ColE9 through the lumen of a single OmpF subunit is driven by the proton-motive force, which is delivered by the TolQ-TolR-TolA-TolB assembly. Our study answers longstanding questions, such as why OmpF is a better translocator than OmpC, and reconciles the mechanisms by which both Tol- and Ton-dependent bacteriocins cross the bacterial outer membrane. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12577.map.gz emd_12577.map.gz | 74.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12577-v30.xml emd-12577-v30.xml emd-12577.xml emd-12577.xml | 24.7 KB 24.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12577_fsc.xml emd_12577_fsc.xml | 13 KB | Display |  FSC data file FSC data file |
| Images |  emd_12577.png emd_12577.png | 18.2 KB | ||
| Masks |  emd_12577_msk_1.map emd_12577_msk_1.map | 83.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12577.cif.gz emd-12577.cif.gz | 7.6 KB | ||
| Others |  emd_12577_additional_1.map.gz emd_12577_additional_1.map.gz emd_12577_half_map_1.map.gz emd_12577_half_map_1.map.gz emd_12577_half_map_2.map.gz emd_12577_half_map_2.map.gz | 41.6 MB 77.8 MB 77.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12577 http://ftp.pdbj.org/pub/emdb/structures/EMD-12577 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12577 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12577 | HTTPS FTP |
-Related structure data
| Related structure data |  7nsuMC  7nstC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12577.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12577.map.gz / Format: CCP4 / Size: 83.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Output map from cryosparc local refinement job, sharpened using deepEMhancer tight target model | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.047 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12577_msk_1.map emd_12577_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened output map from cryosparc local refinement job
| File | emd_12577_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened output map from cryosparc local refinement job | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map A from local refinement job in cryosparc
| File | emd_12577_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A from local refinement job in cryosparc | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B from local refinement job in cryosparc
| File | emd_12577_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B from local refinement job in cryosparc | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ColicinE9 intact translocon complex
| Entire | Name: ColicinE9 intact translocon complex |
|---|---|
| Components |
|
-Supramolecule #1: ColicinE9 intact translocon complex
| Supramolecule | Name: ColicinE9 intact translocon complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Bacteriocin colicinE9 bound to outer membrane protein receptor BtuB and translocator ompF. Disulphide linked to tolB. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 256 KDa |
-Macromolecule #1: Outer membrane protein F
| Macromolecule | Name: Outer membrane protein F / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.11425 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AEIYNKDGNK VDLYGKAVGL HYFSKGNGEN SYGGNGDMTY ARLGFKGETQ INSDLTGYGQ WEYNFQGNNS EGADAQTGNK TRLAFAGLK YADVGSFDYG RNYGVVYDAL GYTDMLPEFG GDTAYSDDFF VGRVGGVATY RNSNFFGLVD GLNFAVQYLG K NERDTARR ...String: AEIYNKDGNK VDLYGKAVGL HYFSKGNGEN SYGGNGDMTY ARLGFKGETQ INSDLTGYGQ WEYNFQGNNS EGADAQTGNK TRLAFAGLK YADVGSFDYG RNYGVVYDAL GYTDMLPEFG GDTAYSDDFF VGRVGGVATY RNSNFFGLVD GLNFAVQYLG K NERDTARR SNGDGVGGSI SYEYEGFGIV GAYGAADRTN LQEAQPLGNG KKAEQWATGL KYDANNIYLA ANYGETRNAT PI TNKFTNT SGFANKTQDV LLVAQYQFDF GLRPSIAYTK SKAKDVEGIG DVDLVNYFEV GATYYFNKNM STYVDYIINQ IDS DNKLGV GSDDTVAVGI VYQF UniProtKB: Outer membrane porin F |
-Macromolecule #2: Colicin-E9
| Macromolecule | Name: Colicin-E9 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 61.707246 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSGGDGRGHN TGAHSTSGNI NGGPTGIGVS GGCSDGSGWS SENNPWGGGS GSGIHWGGGS GRGNGGGNGN SGGGSGTGGN LSAVAAPVA FGFPALSTPG AGGLAVSISA SELSAAIAGI IAKLKKVNLK FTPFGVVLSS LIPSEIAKDD PNMMSKIVTS L PADDITES ...String: MSGGDGRGHN TGAHSTSGNI NGGPTGIGVS GGCSDGSGWS SENNPWGGGS GSGIHWGGGS GRGNGGGNGN SGGGSGTGGN LSAVAAPVA FGFPALSTPG AGGLAVSISA SELSAAIAGI IAKLKKVNLK FTPFGVVLSS LIPSEIAKDD PNMMSKIVTS L PADDITES PVSSLPLDKA TVNVNVRVVD DVKDERQNIS VVSGVPMSVP VVDAKPTERP GVFTASIPGA PVLNISVNDS TP AVQTLSP GVTNNTDKDV RPAGFTQGGN TRDAVIRFPK DSGHNAVYVS VSDVLSPDQV KQRQDEENRR QQEWDATHPV EAA ERNYER ARAELNQANE DVARNQERQA KAVQVYNSRK SELDAANKTL ADAIAEIKQF NRFAHDPMAG GHRMWQMAGL KAQR AQTDV NNKQAAFDAA AKEKSDADAA LSAAQERRKQ KENKEKDAKD KLDKESKRNK PGKATGKGKP VGDKWLDDAG KDSGA PIPD RIADKLRDKE FKSFDDFRKA VWEEVSKDPE LSKNLNPSNK SSVSKGYSPF TPKNQQVGGR KVYELHHDKP ISQGGE VYD MDNIRVTTPK RHIDIHRGK UniProtKB: Colicin-E9 |
-Macromolecule #3: Tol-Pal system protein TolB
| Macromolecule | Name: Tol-Pal system protein TolB / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 46.000285 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKQALRVAFG FLILWASVLH AEVRIVIDSG VDSGRPIGVV PFQWAGPGAA PEDIGGIVAA DLRNSGKFNP LDRARLPQQP GSAQEVQPA AWSALGIDAV VVGQVTPNPD GSYNVAYQLV DTGGAPGTVL AQNSYKVNKQ WLRYAGHTAS DEVFEKLTGI K GAFRTRIA ...String: MKQALRVAFG FLILWASVLH AEVRIVIDSG VDSGRPIGVV PFQWAGPGAA PEDIGGIVAA DLRNSGKFNP LDRARLPQQP GSAQEVQPA AWSALGIDAV VVGQVTPNPD GSYNVAYQLV DTGGAPGTVL AQNSYKVNKQ WLRYAGHTAS DEVFEKLTGI K GAFRTRIA YVVQTNGGQF PYELRVSDYD GYNQFVVHRS PQCLMSPAWS PDGSKLAYVT FESGRSALVI QTLANGAVRQ VA SFPRHNG APAFSPDGSK LAFALSKTGS LNLYVMDLAS GQIRQVTDGR SNNTEPTWFP DSQNLAFTSD QAGRPQVYKV NIN GGAPQR ITWEGSQNQD ADVSSDGKFM VMVSSNGGQQ HIAKQDLATG GVQVLSSTFL DETPSLAPNG TMVIYSSSQG MGSV LNLVS TDGRFKARLP ATDGQVKFPA WSPYL UniProtKB: Tol-Pal system protein TolB |
-Macromolecule #4: Vitamin B12 transporter BtuB
| Macromolecule | Name: Vitamin B12 transporter BtuB / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 66.38618 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QDTSPDTLVV TANRFEQPRS TVLAPTTVVT RQDIDRWQST SVNDVLRRLP GVDITQNGGS GQLSSIFIRG TNASHVLVLI DGVRLNLAG VSGSADLSQF PIALVQRVEY IRGPRSAVYG SDAIGGVVNI ITTRDEPGTE ISAGWGSNSY QNYDVSTQQQ L GDKTRVTL ...String: QDTSPDTLVV TANRFEQPRS TVLAPTTVVT RQDIDRWQST SVNDVLRRLP GVDITQNGGS GQLSSIFIRG TNASHVLVLI DGVRLNLAG VSGSADLSQF PIALVQRVEY IRGPRSAVYG SDAIGGVVNI ITTRDEPGTE ISAGWGSNSY QNYDVSTQQQ L GDKTRVTL LGDYAHTHGY DVVAYGNTGT QAQTDNDGFL SKTLYGALEH NFTDAWSGFV RGYGYDNRTN YDAYYSPGSP LL DTRKLYS QSWDAGLRYN GELIKSQLIT SYSHSKDYNY DPHYGRYDSS ATLDEMKQYT VQWANNVIVG HGSIGAGVDW QKQ TTTPGT GYVEDGYDQR NTGIYLTGLQ QVGDFTFEGA ARSDDNSQFG RHGTWQTSAG WEFIEGYRFI ASYGTSYKAP NLGQ LYGFY GNPNLDPEKS KQWEGAFEGL TAGVNWRISG YRNDVSDLID YDDHTLKYYN EGKARIKGVE ATANFDTGPL THTVS YDYV DARNAITDTP LLRRAKQQVK YQLDWQLYDF DWGITYQYLG TRYDKDYSSY PYQTVKMGGV SLWDLAVAYP VTSHLT VRG KIANLFDKDY ETVYGYQTAG REYTLSGSYT F UniProtKB: Vitamin B12 transporter BtuB |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.9 |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - #0 - Film type ID: 1 / Support film - #0 - Material: GRAPHENE OXIDE / Support film - #0 - topology: CONTINUOUS / Support film - #1 - Film type ID: 2 / Support film - #1 - Material: CARBON / Support film - #1 - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Details: PELCO easiGlow, Ted Pella Inc, USA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 92 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: 3 uL of diluted translocon preparation were applied on freshly glow discharged grids coated with graphene oxide (as described in https://doi.org/10.1038/s41594-019-0355-2); after 30sec ...Details: 3 uL of diluted translocon preparation were applied on freshly glow discharged grids coated with graphene oxide (as described in https://doi.org/10.1038/s41594-019-0355-2); after 30sec waiting grids were blotted for 8-10 using -10 force and plunge frozen in liquid ethane. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 2 / Average exposure time: 10.0 sec. / Average electron dose: 49.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.8000000000000003 µm / Calibrated defocus min: 1.23 µm / Calibrated magnification: 47755 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 47619 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)