+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12434 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mycobacterium smegmatis ATP synthase Fo domain state 1 | |||||||||
 Map data Map data | Ms Fo state1 combiset map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / synthase / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationproton motive force-driven plasma membrane ATP synthesis / proton-transporting two-sector ATPase complex, proton-transporting domain / proton-transporting ATPase activity, rotational mechanism / proton-transporting ATP synthase complex / proton-transporting ATP synthase activity, rotational mechanism / hydrolase activity / lipid binding / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) / Mycolicibacterium smegmatis MC2 155 (bacteria) /  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) / Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) /  Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) | |||||||||
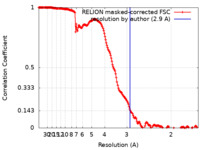
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Montgomery MG / Petri J | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Structure of the ATP synthase from provides targets for treating tuberculosis. Authors: Martin G Montgomery / Jessica Petri / Tobias E Spikes / John E Walker /  Abstract: The structure has been determined by electron cryomicroscopy of the adenosine triphosphate (ATP) synthase from This analysis confirms features in a prior description of the structure of the enzyme, ...The structure has been determined by electron cryomicroscopy of the adenosine triphosphate (ATP) synthase from This analysis confirms features in a prior description of the structure of the enzyme, but it also describes other highly significant attributes not recognized before that are crucial for understanding the mechanism and regulation of the mycobacterial enzyme. First, we resolved not only the three main states in the catalytic cycle described before but also eight substates that portray structural and mechanistic changes occurring during a 360° catalytic cycle. Second, a mechanism of auto-inhibition of ATP hydrolysis involves not only the engagement of the C-terminal region of an α-subunit in a loop in the γ-subunit, as proposed before, but also a "fail-safe" mechanism involving the b'-subunit in the peripheral stalk that enhances engagement. A third unreported characteristic is that the fused bδ-subunit contains a duplicated domain in its N-terminal region where the two copies of the domain participate in similar modes of attachment of the two of three N-terminal regions of the α-subunits. The auto-inhibitory plus the associated "fail-safe" mechanisms and the modes of attachment of the α-subunits provide targets for development of innovative antitubercular drugs. The structure also provides support for an observation made in the bovine ATP synthase that the transmembrane proton-motive force that provides the energy to drive the rotary mechanism is delivered directly and tangentially to the rotor via a Grotthuss water chain in a polar L-shaped tunnel. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12434.map.gz emd_12434.map.gz | 16.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12434-v30.xml emd-12434-v30.xml emd-12434.xml emd-12434.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12434_fsc.xml emd_12434_fsc.xml | 17.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_12434.png emd_12434.png | 92.4 KB | ||
| Filedesc metadata |  emd-12434.cif.gz emd-12434.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12434 http://ftp.pdbj.org/pub/emdb/structures/EMD-12434 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12434 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12434 | HTTPS FTP |
-Validation report
| Summary document |  emd_12434_validation.pdf.gz emd_12434_validation.pdf.gz | 351 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12434_full_validation.pdf.gz emd_12434_full_validation.pdf.gz | 350.6 KB | Display | |
| Data in XML |  emd_12434_validation.xml.gz emd_12434_validation.xml.gz | 15.6 KB | Display | |
| Data in CIF |  emd_12434_validation.cif.gz emd_12434_validation.cif.gz | 21.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12434 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12434 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12434 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12434 | HTTPS FTP |
-Related structure data
| Related structure data |  7nk9MC  7njkC  7njlC  7njmC  7njnC  7njoC  7njpC  7njqC  7njrC  7njsC  7njtC  7njuC  7njvC  7njwC  7njxC  7njyC  7nk7C  7nkbC  7nkdC  7nkhC  7nkjC  7nkkC  7nklC  7nknC  7nkpC  7nkqC  7nl9C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12434.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12434.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Ms Fo state1 combiset map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Mycobacterium smegmatis ATP synthase
| Entire | Name: Mycobacterium smegmatis ATP synthase |
|---|---|
| Components |
|
-Supramolecule #1: Mycobacterium smegmatis ATP synthase
| Supramolecule | Name: Mycobacterium smegmatis ATP synthase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
-Macromolecule #1: ATP synthase gamma chain
| Macromolecule | Name: ATP synthase gamma chain / type: protein_or_peptide / ID: 1 Details: This alignment is incorrect. The initial 13 residues in the model should align with residues 53-65. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 33.439836 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Sequence | String: MAATLRELRG RIRSAGSIKK ITKAQELIAT SRIAKAQARV EAARPYAAEI TNMLTELAGA SALDHPLLVE RKQPKRAGVL VVSSDRGLC GAYNANVLRR AEELFSLLRD EGKDPVLYVV GRKALGYFSF RQRTVVESWT GFSERPTYEN AREIADTLVN A FMAGADDE ...String: MAATLRELRG RIRSAGSIKK ITKAQELIAT SRIAKAQARV EAARPYAAEI TNMLTELAGA SALDHPLLVE RKQPKRAGVL VVSSDRGLC GAYNANVLRR AEELFSLLRD EGKDPVLYVV GRKALGYFSF RQRTVVESWT GFSERPTYEN AREIADTLVN A FMAGADDE GDDAGADGIL GVDELHIVFT EFRSMLSQTA VARRAAPMEV EYVGEVETGP RTLYSFEPDP ETLFDALLPR YI ATRVYAA LLEAAASESA SRRRAMKSAT DNADDLIKAL TLAANRERQA QITQEISEIV GGANALAGSK UniProtKB: ATP synthase gamma chain |
-Macromolecule #2: ATP synthase epsilon chain
| Macromolecule | Name: ATP synthase epsilon chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycobacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 13.277741 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Sequence | String: MADLNVEIVA VERELWSGPA TFVFTRTTAG EIGILPRHIP LVAQLVDDAM VRVEREGEDD LRIAVDGGFL SVTEETVRIL VENAQFESE IDADAAKEDA ASDDERTAAW GRARLRALGQ ID UniProtKB: ATP synthase epsilon chain |
-Macromolecule #3: ATP synthase subunit c
| Macromolecule | Name: ATP synthase subunit c / type: protein_or_peptide / ID: 3 / Details: Uniprot A0R205. The crossref box is missing below. / Number of copies: 9 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 8.597982 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Sequence | String: MDLDPNAIIT AGALIGGGLI MGGGAIGAGI GDGIAGNALI SGIARQPEAQ GRLFTPFFIT VGLVEAAYFI NLAFMALFVF ATPGLQ UniProtKB: ATP synthase subunit c |
-Macromolecule #4: ATP synthase subunit a
| Macromolecule | Name: ATP synthase subunit a / type: protein_or_peptide / ID: 4 / Details: A0R206. The crossref box is missing below. / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 27.568482 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Sequence | String: MLAAEEGGAA IHVGHHTLVF ELFGMTFNGD TILATAVTAV IVIALAFYLR AKVTSTGVPS GVQLFWEALT IQMRQQIEGS IGMKIAPFV LPLSVTIFVF ILISNWLAVL PLQYGGADGA AAELYKAPAS DINFVLALAL FVFVCYHAAG IWRRGIVGHP I KVVKGHVA ...String: MLAAEEGGAA IHVGHHTLVF ELFGMTFNGD TILATAVTAV IVIALAFYLR AKVTSTGVPS GVQLFWEALT IQMRQQIEGS IGMKIAPFV LPLSVTIFVF ILISNWLAVL PLQYGGADGA AAELYKAPAS DINFVLALAL FVFVCYHAAG IWRRGIVGHP I KVVKGHVA FLAPINIVEE LAKPISLALR LFGNIFAGGI LVALIAMFPW YIQWFPNAVW KTFDLFVGLI QAFIFSLLTI LY FSQSMEL DHEDH UniProtKB: ATP synthase subunit a |
-Macromolecule #5: ATP synthase subunit b
| Macromolecule | Name: ATP synthase subunit b / type: protein_or_peptide / ID: 5 / Details: uniprot A0R204. The crossref box is missing below. / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 19.01817 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Sequence | String: MGEFSATILA ASQAAEEGGG GSNFLIPNGT FFAVLIIFLI VLGVISKWVV PPISKVLAER EAMLAKTAAD NRKSAEQVAA AQADYEKEM AEARAQASAL RDEARAAGRS VVDEKRAQAS GEVAQTLTQA DQQLSAQGDQ VRSGLESSVD GLSAKLASRI L GVDVNSGG TQHHHHHHHH HH UniProtKB: ATP synthase subunit b |
-Macromolecule #6: ATP synthase subunit b-delta
| Macromolecule | Name: ATP synthase subunit b-delta / type: protein_or_peptide / ID: 6 / Details: uniprot A0R203. The crossref box is missing. / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria) Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155) (bacteria)Strain: ATCC 700084 / mc(2)155 |
| Molecular weight | Theoretical: 47.504723 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Sequence | String: MSIFIGQLIG FAVIAFIIVK WVVPPVRTLM RNQQEAVRAA LAESAEAAKK LADADAMHAK ALADAKAESE KVTEEAKQDS ERIAAQLSE QAGSEAERIK AQGAQQIQLM RQQLIRQLRT GLGAEAVNKA AEIVRAHVAD PQAQSATVDR FLSELEQMAP S SVVIDTAA ...String: MSIFIGQLIG FAVIAFIIVK WVVPPVRTLM RNQQEAVRAA LAESAEAAKK LADADAMHAK ALADAKAESE KVTEEAKQDS ERIAAQLSE QAGSEAERIK AQGAQQIQLM RQQLIRQLRT GLGAEAVNKA AEIVRAHVAD PQAQSATVDR FLSELEQMAP S SVVIDTAA TSRLRAASRQ SLAALVEKFD SVAGGLDADG LTNLADELAS VAKLLLSETA LNKHLAEPTD DSAPKVRLLE RL LSDKVSA TTLDLLRTAV SNRWSTESNL IDAVEHTARL ALLKRAEIAG EVDEVEEQLF RFGRVLDAEP RLSALLSDYT TPA EGRVAL LDKALTGRPG VNQTAAALLS QTVGLLRGER ADEAVIDLAE LAVSRRGEVV AHVSAAAELS DAQRTRLTEV LSRI YGRPV SVQLHVDPEL LGGLSITVGD EVIDGSIASR LAAAQTGLPD UniProtKB: ATP synthase subunit b-delta |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 59.86 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




































































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)