[English] 日本語
 Yorodumi
Yorodumi- EMDB-1226: Multiple distinct assemblies reveal conformational flexibility in... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1226 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein Hsp26. | |||||||||
 Map data Map data | View down the 2-fold axis of the compact form of yeast Hsp26 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein complex oligomerization / response to salt stress / response to hydrogen peroxide / protein homooligomerization / unfolded protein binding / protein folding / response to heat / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 11.5 Å | |||||||||
 Authors Authors | White HE / Orlova EV / Chen S / Wang L / Ignatiou A / Gowen B / Stromer T / Franzmann TM / Haslbeck M / Buchner J / Saibil HR | |||||||||
 Citation Citation |  Journal: Structure / Year: 2006 Journal: Structure / Year: 2006Title: Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein Hsp26. Authors: Helen E White / Elena V Orlova / Shaoxia Chen / Luchun Wang / Athanasios Ignatiou / Brent Gowen / Thusnelda Stromer / Titus M Franzmann / Martin Haslbeck / Johannes Buchner / Helen R Saibil /  Abstract: Small heat shock proteins are a superfamily of molecular chaperones that suppress protein aggregation and provide protection from cell stress. A key issue for understanding their action is to define ...Small heat shock proteins are a superfamily of molecular chaperones that suppress protein aggregation and provide protection from cell stress. A key issue for understanding their action is to define the interactions of subunit domains in these oligomeric assemblies. Cryo-electron microscopy of yeast Hsp26 reveals two distinct forms, each comprising 24 subunits arranged in a porous shell with tetrahedral symmetry. The subunits form elongated, asymmetric dimers that assemble via trimeric contacts. Modifications of both termini cause rearrangements that yield a further four assemblies. Each subunit contains an N-terminal region, a globular middle domain, the alpha-crystallin domain, and a C-terminal tail. Twelve of the C termini form 3-fold assembly contacts which are inserted into the interior of the shell, while the other 12 C termini form contacts on the surface. Hinge points between the domains allow a variety of assembly contacts, providing the flexibility required for formation of supercomplexes with non-native proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1226.map.gz emd_1226.map.gz | 919.2 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1226-v30.xml emd-1226-v30.xml emd-1226.xml emd-1226.xml | 9.3 KB 9.3 KB | Display Display |  EMDB header EMDB header |
| Images |  1226.gif 1226.gif | 49.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1226 http://ftp.pdbj.org/pub/emdb/structures/EMD-1226 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1226 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1226 | HTTPS FTP |
-Related structure data
| Related structure data |  2h53MC  1221C  1227C  1228C  1229C  1230C  2h50C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1226.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1226.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | View down the 2-fold axis of the compact form of yeast Hsp26 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.86 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
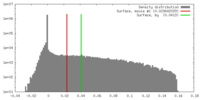
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : yeast small heat shock protein 26
| Entire | Name: yeast small heat shock protein 26 |
|---|---|
| Components |
|
-Supramolecule #1000: yeast small heat shock protein 26
| Supramolecule | Name: yeast small heat shock protein 26 / type: sample / ID: 1000 / Oligomeric state: 24-mer / Number unique components: 1 |
|---|
-Macromolecule #1: Hsp26
| Macromolecule | Name: Hsp26 / type: protein_or_peptide / ID: 1 / Oligomeric state: 24-mer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 40 mM Hepes, 50 mM NaCl, 2 mM EDTA, 1 mM DTT |
| Grid | Details: holey carbon grid |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 1.86 µm / Number real images: 21 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 36080 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.4 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 38000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Phase flipping |
|---|---|
| Final reconstruction | Applied symmetry - Point group: T (tetrahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 11.5 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: Imagic / Details: Angular reconstitution / Number images used: 5000 |
| Final two d classification | Number classes: 770 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name: URO |
| Details | Protocol: Rigid Body. The alpha-crystallin domain dimers were fitted in the program O, prior to refinement. |
| Refinement | Space: RECIPROCAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-2h53: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)