[English] 日本語
 Yorodumi
Yorodumi- EMDB-12078: the 96-nm axonemal repeat from horse sperm flagella (whole population) -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12078 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | the 96-nm axonemal repeat from horse sperm flagella (whole population) | |||||||||
 Map data Map data | subtomogram average of the 96-nm axonemal repeat from horse sperm flagella (whole population) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 40.0 Å | |||||||||
 Authors Authors | Leung MR / Zeev-Ben-Mordehai T | |||||||||
| Funding support |  Netherlands, 1 items Netherlands, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2021 Journal: EMBO J / Year: 2021Title: The multi-scale architecture of mammalian sperm flagella and implications for ciliary motility. Authors: Miguel Ricardo Leung / Marc C Roelofs / Ravi Teja Ravi / Paula Maitan / Heiko Henning / Min Zhang / Elizabeth G Bromfield / Stuart C Howes / Bart M Gadella / Hermes Bloomfield-Gadêlha / ...Authors: Miguel Ricardo Leung / Marc C Roelofs / Ravi Teja Ravi / Paula Maitan / Heiko Henning / Min Zhang / Elizabeth G Bromfield / Stuart C Howes / Bart M Gadella / Hermes Bloomfield-Gadêlha / Tzviya Zeev-Ben-Mordehai /     Abstract: Motile cilia are molecular machines used by a myriad of eukaryotic cells to swim through fluid environments. However, available molecular structures represent only a handful of cell types, limiting ...Motile cilia are molecular machines used by a myriad of eukaryotic cells to swim through fluid environments. However, available molecular structures represent only a handful of cell types, limiting our understanding of how cilia are modified to support motility in diverse media. Here, we use cryo-focused ion beam milling-enabled cryo-electron tomography to image sperm flagella from three mammalian species. We resolve in-cell structures of centrioles, axonemal doublets, central pair apparatus, and endpiece singlets, revealing novel protofilament-bridging microtubule inner proteins throughout the flagellum. We present native structures of the flagellar base, which is crucial for shaping the flagellar beat. We show that outer dense fibers are directly coupled to microtubule doublets in the principal piece but not in the midpiece. Thus, mammalian sperm flagella are ornamented across scales, from protofilament-bracing structures reinforcing microtubules at the nano-scale to accessory structures that impose micron-scale asymmetries on the entire assembly. Our structures provide vital foundations for linking molecular structure to ciliary motility and evolution. #1:  Journal: Biorxiv / Year: 2020 Journal: Biorxiv / Year: 2020Title: The multi-scale architecture of mammalian sperm flagella and implications for ciliary motility Authors: Leung MR / Roelofs MC / Ravi RT / Maitan P / Zhang M / Henning H / Bromfield EG / Howes SC / Gadella BM / Bloomfield-Gadelha H / Zeev-Ben-Mordehai T | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12078.map.gz emd_12078.map.gz | 3.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12078-v30.xml emd-12078-v30.xml emd-12078.xml emd-12078.xml | 10.1 KB 10.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_12078.png emd_12078.png | 157.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12078 http://ftp.pdbj.org/pub/emdb/structures/EMD-12078 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12078 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12078 | HTTPS FTP |
-Validation report
| Summary document |  emd_12078_validation.pdf.gz emd_12078_validation.pdf.gz | 277.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12078_full_validation.pdf.gz emd_12078_full_validation.pdf.gz | 276.8 KB | Display | |
| Data in XML |  emd_12078_validation.xml.gz emd_12078_validation.xml.gz | 5.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12078 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12078 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12078 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12078 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12078.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12078.map.gz / Format: CCP4 / Size: 3.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | subtomogram average of the 96-nm axonemal repeat from horse sperm flagella (whole population) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 11.32 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
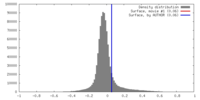
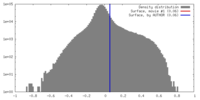
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : subtomogram average of the 96-nm axonemal repeat from horse sperm...
| Entire | Name: subtomogram average of the 96-nm axonemal repeat from horse sperm flagella (whole population) |
|---|---|
| Components |
|
-Supramolecule #1: subtomogram average of the 96-nm axonemal repeat from horse sperm...
| Supramolecule | Name: subtomogram average of the 96-nm axonemal repeat from horse sperm flagella (whole population) type: cell / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 1.67 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 40.0 Å / Resolution method: FSC 0.5 CUT-OFF / Number subtomograms used: 634 |
|---|---|
| Extraction | Number tomograms: 16 / Number images used: 634 / Software - Name: PEET (ver. 1.13.0) |
| CTF correction | Software - Name: PEET (ver. 1.13.0) |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)