+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12064 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Yeast 80S ribosome bound to eEF3 and P/P-tRNA (POST-3) | ||||||||||||||||||
 Map data Map data | Cryo-EM structure of the S. cerevisiae 80S ribosome in complex with eEF3 and P/P-site tRNA (postprocessed masked) | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
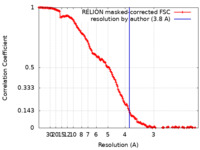
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | ||||||||||||||||||
 Authors Authors | Ranjan N / Pochopien AA / Wu CC / Beckert B / Blanchet S / Green R / Rodnina MV / Wilson DN | ||||||||||||||||||
| Funding support | 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: EMBO J / Year: 2021 Journal: EMBO J / Year: 2021Title: Yeast translation elongation factor eEF3 promotes late stages of tRNA translocation. Authors: Namit Ranjan / Agnieszka A Pochopien / Colin Chih-Chien Wu / Bertrand Beckert / Sandra Blanchet / Rachel Green / Marina V Rodnina / Daniel N Wilson /   Abstract: In addition to the conserved translation elongation factors eEF1A and eEF2, fungi require a third essential elongation factor, eEF3. While eEF3 has been implicated in tRNA binding and release at the ...In addition to the conserved translation elongation factors eEF1A and eEF2, fungi require a third essential elongation factor, eEF3. While eEF3 has been implicated in tRNA binding and release at the ribosomal A and E sites, its exact mechanism of action is unclear. Here, we show that eEF3 acts at the mRNA-tRNA translocation step by promoting the dissociation of the tRNA from the E site, but independent of aminoacyl-tRNA recruitment to the A site. Depletion of eEF3 in vivo leads to a general slowdown in translation elongation due to accumulation of ribosomes with an occupied A site. Cryo-EM analysis of native eEF3-ribosome complexes shows that eEF3 facilitates late steps of translocation by favoring non-rotated ribosomal states, as well as by opening the L1 stalk to release the E-site tRNA. Additionally, our analysis provides structural insights into novel translation elongation states, enabling presentation of a revised yeast translation elongation cycle. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12064.map.gz emd_12064.map.gz | 21.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12064-v30.xml emd-12064-v30.xml emd-12064.xml emd-12064.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12064_fsc.xml emd_12064_fsc.xml | 14.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_12064.png emd_12064.png | 199.7 KB | ||
| Others |  emd_12064_additional_1.map.gz emd_12064_additional_1.map.gz emd_12064_half_map_1.map.gz emd_12064_half_map_1.map.gz emd_12064_half_map_2.map.gz emd_12064_half_map_2.map.gz | 224.8 MB 225.7 MB 225.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12064 http://ftp.pdbj.org/pub/emdb/structures/EMD-12064 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12064 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12064 | HTTPS FTP |
-Validation report
| Summary document |  emd_12064_validation.pdf.gz emd_12064_validation.pdf.gz | 442.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12064_full_validation.pdf.gz emd_12064_full_validation.pdf.gz | 441.6 KB | Display | |
| Data in XML |  emd_12064_validation.xml.gz emd_12064_validation.xml.gz | 23 KB | Display | |
| Data in CIF |  emd_12064_validation.cif.gz emd_12064_validation.cif.gz | 29.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12064 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12064 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12064 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12064 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12064.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12064.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the S. cerevisiae 80S ribosome in complex with eEF3 and P/P-site tRNA (postprocessed masked) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.084 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Cryo-EM structure of the S. cerevisiae 80S ribosome...
| File | emd_12064_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the S. cerevisiae 80S ribosome in complex with eEF3 and P/P-site tRNA (refinement) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM structure of the S. cerevisiae 80S ribosome...
| File | emd_12064_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the S. cerevisiae 80S ribosome in complex with eEF3 and P/P-site tRNA (half map 1) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Cryo-EM structure of the S. cerevisiae 80S ribosome...
| File | emd_12064_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of the S. cerevisiae 80S ribosome in complex with eEF3 and P/P-site tRNA (half map 2) | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the S. cerevisiae 80S ribosome in complex wi...
| Entire | Name: Cryo-EM structure of the S. cerevisiae 80S ribosome in complex with eEF3 and A/A- and P/P-site tRNAs. |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the S. cerevisiae 80S ribosome in complex wi...
| Supramolecule | Name: Cryo-EM structure of the S. cerevisiae 80S ribosome in complex with eEF3 and A/A- and P/P-site tRNAs. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z
Z Y
Y X
X