+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11971 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
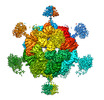
| Title | Stressosome complex from Listeria innocua | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Stressosome / stress sensing / general stress response / phosphorylation cascade / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Listeria innocua Clip11262 (bacteria) / Listeria innocua Clip11262 (bacteria) /  Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) (bacteria) Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.86 Å | |||||||||
 Authors Authors | Miksys A / Fu L | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2022 Journal: Commun Biol / Year: 2022Title: Molecular insights into intra-complex signal transmission during stressosome activation. Authors: Algirdas Miksys / Lifei Fu / M Gregor Madej / Duarte N Guerreiro / Susann Kaltwasser / Maria Conway / Sema Ejder / Astrid Bruckmann / Jon Marles-Wright / Richard J Lewis / Conor O'Byrne / ...Authors: Algirdas Miksys / Lifei Fu / M Gregor Madej / Duarte N Guerreiro / Susann Kaltwasser / Maria Conway / Sema Ejder / Astrid Bruckmann / Jon Marles-Wright / Richard J Lewis / Conor O'Byrne / Jan Pané-Farré / Christine Ziegler /    Abstract: The stressosome is a pseudo-icosahedral megadalton bacterial stress-sensing protein complex consisting of several copies of two STAS-domain proteins, RsbR and RsbS, and the kinase RsbT. Upon ...The stressosome is a pseudo-icosahedral megadalton bacterial stress-sensing protein complex consisting of several copies of two STAS-domain proteins, RsbR and RsbS, and the kinase RsbT. Upon perception of environmental stress multiple copies of RsbT are released from the surface of the stressosome. Free RsbT activates downstream proteins to elicit a global cellular response, such as the activation of the general stress response in Gram-positive bacteria. The molecular events triggering RsbT release from the stressosome surface remain poorly understood. Here we present the map of Listeria innocua RsbR1/RsbS complex at resolutions of 3.45 Å for the STAS domain core in icosahedral symmetry and of 3.87 Å for the STAS domain and N-terminal sensors in D2 symmetry, respectively. The structure reveals a conformational change in the STAS domain linked to phosphorylation in RsbR. Docking studies indicate that allosteric RsbT binding to the conformationally flexible N-terminal sensor domain of RsbR affects the affinity of RsbS towards RsbT. Our results bring to focus the molecular events within the stressosome complex and further our understanding of this ubiquitous signaling hub. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11971.map.gz emd_11971.map.gz | 200.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11971-v30.xml emd-11971-v30.xml emd-11971.xml emd-11971.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_11971.png emd_11971.png | 167.5 KB | ||
| Filedesc metadata |  emd-11971.cif.gz emd-11971.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11971 http://ftp.pdbj.org/pub/emdb/structures/EMD-11971 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11971 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11971 | HTTPS FTP |
-Related structure data
| Related structure data |  7b0uMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11971.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11971.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0635 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : stressosome complex RsbR and RsbS
| Entire | Name: stressosome complex RsbR and RsbS |
|---|---|
| Components |
|
-Supramolecule #1: stressosome complex RsbR and RsbS
| Supramolecule | Name: stressosome complex RsbR and RsbS / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Listeria innocua Clip11262 (bacteria) Listeria innocua Clip11262 (bacteria) |
| Molecular weight | Theoretical: 1.5 MDa |
-Macromolecule #1: RsbR protein
| Macromolecule | Name: RsbR protein / type: protein_or_peptide / ID: 1 / Number of copies: 16 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) (bacteria) Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) (bacteria)Strain: ATCC BAA-680 / CLIP 11262 |
| Molecular weight | Theoretical: 31.704586 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MYKDFANFIR TNKKDLLNNW MNEMEKQSDP LINDIAKEPM YEETSIEFVD LIVSNITENG SKFNEKLDDF AEKVVHLGWP IHFVTTGLR VFGLLVYTAM RDEDLFLKRE EKPEDDAYYR FETWLSSMYN KVVTAYADTW EKTVSIQKSA LQELSAPLLP I FEKISVMP ...String: MYKDFANFIR TNKKDLLNNW MNEMEKQSDP LINDIAKEPM YEETSIEFVD LIVSNITENG SKFNEKLDDF AEKVVHLGWP IHFVTTGLR VFGLLVYTAM RDEDLFLKRE EKPEDDAYYR FETWLSSMYN KVVTAYADTW EKTVSIQKSA LQELSAPLLP I FEKISVMP LIGTIDTERA KLIIENLLIG VVKNRSEVVL IDITGVPVVD TMVAHHIIQA SEAVRLVGCQ AMLVGIRPEI AQ TIVNLGI ELDQIITTNT MKKGMERALA LTNREIVEKE G UniProtKB: RsbR protein |
-Macromolecule #2: RsbR protein
| Macromolecule | Name: RsbR protein / type: protein_or_peptide / ID: 2 / Number of copies: 16 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) (bacteria) Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) (bacteria)Strain: ATCC BAA-680 / CLIP 11262 |
| Molecular weight | Theoretical: 31.784566 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MYKDFANFIR TNKKDLLNNW MNEMEKQSDP LINDIAKEPM YEETSIEFVD LIVSNITENG SKFNEKLDDF AEKVVHLGWP IHFVTTGLR VFGLLVYTAM RDEDLFLKRE EKPEDDAYYR FETWLSSMYN KVVTAYADTW EKTVSIQKSA LQELSAPLLP I FEKISVMP ...String: MYKDFANFIR TNKKDLLNNW MNEMEKQSDP LINDIAKEPM YEETSIEFVD LIVSNITENG SKFNEKLDDF AEKVVHLGWP IHFVTTGLR VFGLLVYTAM RDEDLFLKRE EKPEDDAYYR FETWLSSMYN KVVTAYADTW EKTVSIQKSA LQELSAPLLP I FEKISVMP LIGTIDTERA KLIIENLLIG VVKNRSEVVL IDITGVPVVD TMVAHHIIQA SEAVRLVGCQ AMLVGIRPEI AQ (TPO)IVNLGI ELDQIITTNT MKKGMERALA LTNREIVEKE G UniProtKB: RsbR protein |
-Macromolecule #3: RsbS protein
| Macromolecule | Name: RsbS protein / type: protein_or_peptide / ID: 3 / Number of copies: 20 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) (bacteria) Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) (bacteria)Strain: ATCC BAA-680 / CLIP 11262 |
| Molecular weight | Theoretical: 12.606707 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGIPILKLGE CLLISIQSEL DDHTAVEFQE DLLAKIHETS ARGVVIDITS IDFIDSFIAK ILGDVVSMSK LMGAKVVVTG IQPAVAITL IELGITFSGV LSAMDLESGL EKLKQELGE UniProtKB: RsbS protein |
-Macromolecule #4: RsbR protein
| Macromolecule | Name: RsbR protein / type: protein_or_peptide / ID: 4 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) (bacteria) Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) (bacteria)Strain: ATCC BAA-680 / CLIP 11262 |
| Molecular weight | Theoretical: 31.864547 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MYKDFANFIR TNKKDLLNNW MNEMEKQSDP LINDIAKEPM YEETSIEFVD LIVSNITENG SKFNEKLDDF AEKVVHLGWP IHFVTTGLR VFGLLVYTAM RDEDLFLKRE EKPEDDAYYR FETWLSSMYN KVVTAYADTW EKTVSIQKSA LQELSAPLLP I FEKISVMP ...String: MYKDFANFIR TNKKDLLNNW MNEMEKQSDP LINDIAKEPM YEETSIEFVD LIVSNITENG SKFNEKLDDF AEKVVHLGWP IHFVTTGLR VFGLLVYTAM RDEDLFLKRE EKPEDDAYYR FETWLSSMYN KVVTAYADTW EKTVSIQKSA LQELSAPLLP I FEKISVMP LIGTID(TPO)ERA KLIIENLLIG VVKNRSEVVL IDITGVPVVD TMVAHHIIQA SEAVRLVGCQ AMLVGIRP E IAQ(TPO)IVNLGI ELDQIITTNT MKKGMERALA LTNREIVEKE G UniProtKB: RsbR protein |
-Macromolecule #5: RsbR protein
| Macromolecule | Name: RsbR protein / type: protein_or_peptide / ID: 5 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) (bacteria) Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) (bacteria)Strain: ATCC BAA-680 / CLIP 11262 |
| Molecular weight | Theoretical: 31.784566 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MYKDFANFIR TNKKDLLNNW MNEMEKQSDP LINDIAKEPM YEETSIEFVD LIVSNITENG SKFNEKLDDF AEKVVHLGWP IHFVTTGLR VFGLLVYTAM RDEDLFLKRE EKPEDDAYYR FETWLSSMYN KVVTAYADTW EKTVSIQKSA LQELSAPLLP I FEKISVMP ...String: MYKDFANFIR TNKKDLLNNW MNEMEKQSDP LINDIAKEPM YEETSIEFVD LIVSNITENG SKFNEKLDDF AEKVVHLGWP IHFVTTGLR VFGLLVYTAM RDEDLFLKRE EKPEDDAYYR FETWLSSMYN KVVTAYADTW EKTVSIQKSA LQELSAPLLP I FEKISVMP LIGTID(TPO)ERA KLIIENLLIG VVKNRSEVVL IDITGVPVVD TMVAHHIIQA SEAVRLVGCQ AMLVGIRP E IAQTIVNLGI ELDQIITTNT MKKGMERALA LTNREIVEKE G UniProtKB: RsbR protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8.5 Component:
Details: pH 8.5 | |||||||||
| Grid | Model: Quantifoil / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 120 sec. / Pretreatment - Pressure: 0.029 kPa | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Applied symmetry - Point group: D2 (2x2 fold dihedral) / Resolution.type: BY AUTHOR / Resolution: 3.86 Å / Resolution method: FSC 0.143 CUT-OFF / Software: (Name: RELION (ver. 3.0), cryoSPARC (ver. V2)) / Number images used: 32031 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-7b0u: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)





















