+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-1197 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. | |||||||||
 マップデータ マップデータ | 3D reconstruction of IBV pseudoknot-stalled rabbit reticulocyte ribosome containing tRNA, eEF2, pseudoknot and both ribosomal subunits. | |||||||||
 試料 試料 |
| |||||||||
| 生物種 |  Infectious bronchitis virus (伝染性気管支炎ウイルス) / Infectious bronchitis virus (伝染性気管支炎ウイルス) /  | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 16.2 Å | |||||||||
 データ登録者 データ登録者 | Namy O / Moran SJ / Stuart DI / Gilbert RJC / Brierley I | |||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2006 ジャーナル: Nature / 年: 2006タイトル: A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. 著者: Olivier Namy / Stephen J Moran / David I Stuart / Robert J C Gilbert / Ian Brierley /  要旨: The triplet-based genetic code requires that translating ribosomes maintain the reading frame of a messenger RNA faithfully to ensure correct protein synthesis. However, in programmed -1 ribosomal ...The triplet-based genetic code requires that translating ribosomes maintain the reading frame of a messenger RNA faithfully to ensure correct protein synthesis. However, in programmed -1 ribosomal frameshifting, a specific subversion of frame maintenance takes place, wherein the ribosome is forced to shift one nucleotide backwards into an overlapping reading frame and to translate an entirely new sequence of amino acids. This process is indispensable in the replication of numerous viral pathogens, including HIV and the coronavirus associated with severe acute respiratory syndrome, and is also exploited in the expression of several cellular genes. Frameshifting is promoted by an mRNA signal composed of two essential elements: a heptanucleotide 'slippery' sequence and an adjacent mRNA secondary structure, most often an mRNA pseudoknot. How these components operate together to manipulate the ribosome is unknown. Here we describe the observation of a ribosome-mRNA pseudoknot complex that is stalled in the process of -1 frameshifting. Cryoelectron microscopic imaging of purified mammalian 80S ribosomes from rabbit reticulocytes paused at a coronavirus pseudoknot reveals an intermediate of the frameshifting process. From this it can be seen how the pseudoknot interacts with the ribosome to block the mRNA entrance channel, compromising the translocation process and leading to a spring-like deformation of the P-site transfer RNA. In addition, we identify movements of the likely eukaryotic ribosomal helicase and confirm a direct interaction between the translocase eEF2 and the P-site tRNA. Together, the structural changes provide a mechanical explanation of how the pseudoknot manipulates the ribosome into a different reading frame. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_1197.map.gz emd_1197.map.gz | 494.9 KB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-1197-v30.xml emd-1197-v30.xml emd-1197.xml emd-1197.xml | 11.1 KB 11.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  1197.gif 1197.gif | 45.3 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1197 http://ftp.pdbj.org/pub/emdb/structures/EMD-1197 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1197 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1197 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_1197_validation.pdf.gz emd_1197_validation.pdf.gz | 220.3 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_1197_full_validation.pdf.gz emd_1197_full_validation.pdf.gz | 219.4 KB | 表示 | |
| XML形式データ |  emd_1197_validation.xml.gz emd_1197_validation.xml.gz | 5.9 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1197 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1197 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1197 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-1197 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_1197.map.gz / 形式: CCP4 / 大きさ: 15.3 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_1197.map.gz / 形式: CCP4 / 大きさ: 15.3 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | 3D reconstruction of IBV pseudoknot-stalled rabbit reticulocyte ribosome containing tRNA, eEF2, pseudoknot and both ribosomal subunits. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 3.33 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
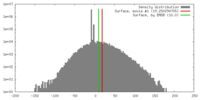
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : Rabbit reticuloyte ribosome stalled on IBV mRNA containing pseudo...
| 全体 | 名称: Rabbit reticuloyte ribosome stalled on IBV mRNA containing pseudoknot with tRNA and eEF2 |
|---|---|
| 要素 |
|
-超分子 #1000: Rabbit reticuloyte ribosome stalled on IBV mRNA containing pseudo...
| 超分子 | 名称: Rabbit reticuloyte ribosome stalled on IBV mRNA containing pseudoknot with tRNA and eEF2 タイプ: sample / ID: 1000 集合状態: One each of 40S, 60S, mRNA, eEF2 and P-site tRNA Number unique components: 5 |
|---|---|
| 分子量 | 実験値: 3.5 MDa / 理論値: 3.5 MDa |
-超分子 #1: Small subunit
| 超分子 | 名称: Small subunit / タイプ: complex / ID: 1 / Name.synonym: 40S / Ribosome-details: ribosome-eukaryote: SSU 40S |
|---|
-超分子 #2: Large subunit
| 超分子 | 名称: Large subunit / タイプ: complex / ID: 2 / Name.synonym: 60S / Ribosome-details: ribosome-eukaryote: LSU 60S |
|---|
-分子 #1: Message with pseudoknot
| 分子 | 名称: Message with pseudoknot / タイプ: rna / ID: 1 / Name.synonym: mRNA / 分類: OTHER / Structure: SINGLE STRANDED / Synthetic?: No |
|---|---|
| 由来(天然) | 生物種:  Infectious bronchitis virus (伝染性気管支炎ウイルス) Infectious bronchitis virus (伝染性気管支炎ウイルス)別称: IBV |
-分子 #3: P-site transfer RNA
| 分子 | 名称: P-site transfer RNA / タイプ: rna / ID: 3 / Name.synonym: tRNA / 分類: OTHER / Structure: DOUBLE HELIX / Synthetic?: No |
|---|---|
| 由来(天然) | 生物種:  |
-分子 #2: Elongation translocase
| 分子 | 名称: Elongation translocase / タイプ: protein_or_peptide / ID: 2 / Name.synonym: eEF2 / コピー数: 1 / 集合状態: Monomer / 組換発現: No |
|---|---|
| 由来(天然) | 生物種:  |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| グリッド | 詳細: 300 mesh gold w/ lacey carbon |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI/PHILIPS CM200FEG |
|---|---|
| 温度 | 平均: 100 K |
| 撮影 | カテゴリ: FILM / フィルム・検出器のモデル: KODAK SO-163 FILM / デジタル化 - スキャナー: OTHER / デジタル化 - サンプリング間隔: 3.33 µm / 実像数: 31 / 平均電子線量: 2 e/Å2 / Od range: 5 / ビット/ピクセル: 8 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: SPOT SCAN / 撮影モード: BRIGHT FIELD / Cs: 2 mm / 最大 デフォーカス(公称値): 7.0 µm / 最小 デフォーカス(公称値): 1.0 µm / 倍率(公称値): 50000 |
| 試料ステージ | 試料ホルダー: Eucentric / 試料ホルダーモデル: GATAN LIQUID NITROGEN |
- 画像解析
画像解析
| 詳細 | The particles were selected in a semi-automated fashion using BOXER (EMAN suite) |
|---|---|
| CTF補正 | 詳細: By micrograph |
| 最終 再構成 | 想定した対称性 - 点群: C1 (非対称) / アルゴリズム: OTHER / 解像度のタイプ: BY AUTHOR / 解像度: 16.2 Å / 解像度の算出法: FSC 0.5 CUT-OFF / ソフトウェア - 名称: SPIDER and GAP 詳細: Deposited map is composite of computationally-separated subunits and bound co-factors from the final map. 使用した粒子像数: 17672 |
| 最終 角度割当 | 詳細: SPIDER |
-原子モデル構築 1
| ソフトウェア | 名称: URO |
|---|---|
| 詳細 | Protocol: Rigid body. Domains manually docked in O and refined in URO |
| 精密化 | プロトコル: RIGID BODY FIT / 当てはまり具合の基準: CC |
 ムービー
ムービー コントローラー
コントローラー











 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)