[English] 日本語
 Yorodumi
Yorodumi- EMDB-11813: Structure of the SARS-CoV spike glycoprotein in complex with the ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11813 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
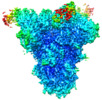
| Title | Structure of the SARS-CoV spike glycoprotein in complex with the 47D11 neutralizing antibody Fab fragment | ||||||||||||||||||
 Map data Map data | Complex of the SARS-CoV spike glycoprotein with the neutralizing antibody Fab fragment 47D11 | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | SARS-coV / spike / neutralizing antibody / VIRAL PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane ...host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / identical protein binding / membrane Similarity search - Function | ||||||||||||||||||
| Biological species |   SARS coronavirus WH20 / SARS coronavirus WH20 /  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | ||||||||||||||||||
 Authors Authors | Fedry J / Hurdiss DL | ||||||||||||||||||
| Funding support |  Netherlands, 5 items Netherlands, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: Structural insights into the cross-neutralization of SARS-CoV and SARS-CoV-2 by the human monoclonal antibody 47D11. Authors: Juliette Fedry / Daniel L Hurdiss / Chunyan Wang / Wentao Li / Gonzalo Obal / Ieva Drulyte / Wenjuan Du / Stuart C Howes / Frank J M van Kuppeveld / Friedrich Förster / Berend-Jan Bosch /  Abstract: The emergence of SARS-CoV-2 antibody escape mutations highlights the urgent need for broadly neutralizing therapeutics. We previously identified a human monoclonal antibody, 47D11, capable of cross- ...The emergence of SARS-CoV-2 antibody escape mutations highlights the urgent need for broadly neutralizing therapeutics. We previously identified a human monoclonal antibody, 47D11, capable of cross-neutralizing SARS-CoV-2 and SARS-CoV and protecting against the associated respiratory disease in an animal model. Here, we report cryo-EM structures of both trimeric spike ectodomains in complex with the 47D11 Fab. 47D11 binds to the closed receptor-binding domain, distal to the ACE2 binding site. The CDRL3 stabilizes the N343 glycan in an upright conformation, exposing a mutationally constrained hydrophobic pocket, into which the CDRH3 loop inserts two aromatic residues. 47D11 stabilizes a partially open conformation of the SARS-CoV-2 spike, suggesting that it could be used effectively in combination with other antibodies targeting the exposed receptor-binding motif. Together, these results reveal a cross-protective epitope on the SARS-CoV-2 spike and provide a structural roadmap for the development of 47D11 as a prophylactic or postexposure therapy for COVID-19. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11813.map.gz emd_11813.map.gz | 23.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11813-v30.xml emd-11813-v30.xml emd-11813.xml emd-11813.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11813_fsc.xml emd_11813_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_11813.png emd_11813.png | 212.4 KB | ||
| Filedesc metadata |  emd-11813.cif.gz emd-11813.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11813 http://ftp.pdbj.org/pub/emdb/structures/EMD-11813 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11813 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11813 | HTTPS FTP |
-Related structure data
| Related structure data |  7akjMC  7akdC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11813.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11813.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Complex of the SARS-CoV spike glycoprotein with the neutralizing antibody Fab fragment 47D11 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of the SARS-CoV-2 spike glycoprotein with the 47D11 neutr...
| Entire | Name: Complex of the SARS-CoV-2 spike glycoprotein with the 47D11 neutralizing antibody Fab fragment |
|---|---|
| Components |
|
-Supramolecule #1: Complex of the SARS-CoV-2 spike glycoprotein with the 47D11 neutr...
| Supramolecule | Name: Complex of the SARS-CoV-2 spike glycoprotein with the 47D11 neutralizing antibody Fab fragment type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 600 KDa |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  SARS coronavirus WH20 SARS coronavirus WH20 |
| Molecular weight | Theoretical: 132.302688 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: RCTTFDDVQA PNYTQHTSSM RGVYYPDEIF RSDTLYLTQD LFLPFYSNVT GFHTINHTFD NPVIPFKDGI YFAATEKSNV VRGWVFGST MNNKSQSVII INNSTNVVIR ACNFELCDNP FFAVSKPMGT QTHTMIFDNA FNCTFEYISD AFSLDVSEKS G NFKHLREF ...String: RCTTFDDVQA PNYTQHTSSM RGVYYPDEIF RSDTLYLTQD LFLPFYSNVT GFHTINHTFD NPVIPFKDGI YFAATEKSNV VRGWVFGST MNNKSQSVII INNSTNVVIR ACNFELCDNP FFAVSKPMGT QTHTMIFDNA FNCTFEYISD AFSLDVSEKS G NFKHLREF VFKNKDGFLY VYKGYQPIDV VRDLPSGFNT LKPIFKLPLG INITNFRAIL TAFSPAQDTW GTSAAAYFVG YL KPTTFML KYDENGTITD AVDCSQNPLA ELKCSVKSFE IDKGIYQTSN FRVVPSGDVV RFPNITNLCP FGEVFNATKF PSV YAWERK KISNCVADYS VLYNSTFFST FKCYGVSATK LNDLCFSNVY ADSFVVKGDD VRQIAPGQTG VIADYNYKLP DDFM GCVLA WNTRNIDATS TGNYNYKYRY LRHGKLRPFE RDISNVPFSP DGKPCTPPAL NCYWPLNDYG FYTTTGIGYQ PYRVV VLSF ELLNAPATVC GPKLSTDLIK NQCVNFNFNG LTGTGVLTPS SKRFQPFQQF GRDVSDFTDS VRDPKTSEIL DISPCS FGG VSVITPGTNA SSEVAVLYQD VNCTDVSTAI HADQLTPAWR IYSTGNNVFQ TQAGCLIGAE HVDTSYECDI PIGAGIC AS YHTVSLLRST SQKSIVAYTM SLGADSSIAY SNNTIAIPTN FSISITTEVM PVSMAKTSVD CNMYICGDST ECANLLLQ Y GSFCTQLNRA LSGIAAEQDR NTREVFAQVK QMYKTPTLKY FGGFNFSQIL PDPLKPTKRS FIEDLLFNKV TLADAGFMK QYGECLGDIN ARDLICAQKF NGLTVLPPLL TDDMIAAYTA ALVSGTATAG WTFGAGAALQ IPFAMQMAYR FNGIGVTQNV LYENQKQIA NQFNKAISQI QESLTTTSTA LGKLQDVVNQ NAQALNTLVK QLSSNFGAIS SVLNDILSRL DPPEAEVQID R LITGRLQS LQTYVTQQLI RAAEIRASAN LAATKMSECV LGQSKRVDFC GKGYHLMSFP QAAPHGVVFL HVTYVPSQER NF TTAPAIC HEGKAYFPRE GVFVFNGTSW FITQRNFFSP QIITTDNTFV SGNCDVVIGI INNTVYDPLQ PELDSFKEEL DKY FKNHTS PDVDLGDISG INASVVNLIK GSGYIPEAPR DGQAYVRKDG EWVLLSTFLI KLVPRGSLEW SHPQFEK UniProtKB: Spike glycoprotein |
-Macromolecule #2: 47D11 neutralizing antibody heavy chain
| Macromolecule | Name: 47D11 neutralizing antibody heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.205812 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLQESGPG LVKPSETLSL TCSVSGGSIS SHYWSWIRQP PGKGLEWIGY IYYSGSTNHN PSLKSRVTIS VDTSKNQFSL KLSSVTAAD TAVYYCARGV LLWFGEPIFE IWGQGTMVTV SS |
-Macromolecule #3: 47D11 neutralizing antibody light chain
| Macromolecule | Name: 47D11 neutralizing antibody light chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.350607 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVMTQSPAT LSVSPGERAT LSCRASQSVS SSLAWYQQKP GQAPRLLIYG ASTRAPGIPA RFSGSGSGTE FTLTISSLQS EDFAVYYCQ QYNNWPLTFG GGTKVEI |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 36 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.6 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 4231 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 30.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)























