[English] 日本語
 Yorodumi
Yorodumi- EMDB-11657: CryoEM structure of a human gamma-aminobutyric acid receptor, the... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11657 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of a human gamma-aminobutyric acid receptor, the GABA(A)R-beta3 homopentamer, in complex with histamine and megabody Mb25 in lipid nanodisc | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Pentameric ligand-gated ion channel / Neurotrasmitter receptor / GABA(A) receptor / MEMBRANE PROTEIN | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcircadian sleep/wake cycle, REM sleep / reproductive behavior / hard palate development / cellular response to histamine / GABA receptor activation / inner ear receptor cell development / inhibitory synapse assembly / GABA-A receptor activity / GABA-gated chloride ion channel activity / GABA-A receptor complex ...circadian sleep/wake cycle, REM sleep / reproductive behavior / hard palate development / cellular response to histamine / GABA receptor activation / inner ear receptor cell development / inhibitory synapse assembly / GABA-A receptor activity / GABA-gated chloride ion channel activity / GABA-A receptor complex / innervation / response to anesthetic / postsynaptic specialization membrane / inhibitory postsynaptic potential / gamma-aminobutyric acid signaling pathway / synaptic transmission, GABAergic / cellular response to zinc ion / exploration behavior / motor behavior / roof of mouth development / Signaling by ERBB4 / cochlea development / social behavior / chloride channel complex / cytoplasmic vesicle membrane / chloride transmembrane transport / cerebellum development / learning / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / GABA-ergic synapse / memory / dendritic spine / postsynaptic membrane / response to xenobiotic stimulus / cell surface / signal transduction / identical protein binding / plasma membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||||||||||||||
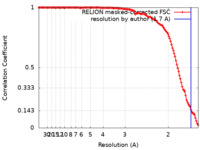
| Method | single particle reconstruction / cryo EM / Resolution: 1.7 Å | |||||||||||||||||||||
 Authors Authors | Nakane T / Kotecha A | |||||||||||||||||||||
| Funding support |  United Kingdom, 6 items United Kingdom, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Single-particle cryo-EM at atomic resolution. Authors: Takanori Nakane / Abhay Kotecha / Andrija Sente / Greg McMullan / Simonas Masiulis / Patricia M G E Brown / Ioana T Grigoras / Lina Malinauskaite / Tomas Malinauskas / Jonas Miehling / ...Authors: Takanori Nakane / Abhay Kotecha / Andrija Sente / Greg McMullan / Simonas Masiulis / Patricia M G E Brown / Ioana T Grigoras / Lina Malinauskaite / Tomas Malinauskas / Jonas Miehling / Tomasz Uchański / Lingbo Yu / Dimple Karia / Evgeniya V Pechnikova / Erwin de Jong / Jeroen Keizer / Maarten Bischoff / Jamie McCormack / Peter Tiemeijer / Steven W Hardwick / Dimitri Y Chirgadze / Garib Murshudov / A Radu Aricescu / Sjors H W Scheres /    Abstract: The three-dimensional positions of atoms in protein molecules define their structure and their roles in biological processes. The more precisely atomic coordinates are determined, the more chemical ...The three-dimensional positions of atoms in protein molecules define their structure and their roles in biological processes. The more precisely atomic coordinates are determined, the more chemical information can be derived and the more mechanistic insights into protein function may be inferred. Electron cryo-microscopy (cryo-EM) single-particle analysis has yielded protein structures with increasing levels of detail in recent years. However, it has proved difficult to obtain cryo-EM reconstructions with sufficient resolution to visualize individual atoms in proteins. Here we use a new electron source, energy filter and camera to obtain a 1.7 Å resolution cryo-EM reconstruction for a human membrane protein, the β3 GABA receptor homopentamer. Such maps allow a detailed understanding of small-molecule coordination, visualization of solvent molecules and alternative conformations for multiple amino acids, and unambiguous building of ordered acidic side chains and glycans. Applied to mouse apoferritin, our strategy led to a 1.22 Å resolution reconstruction that offers a genuine atomic-resolution view of a protein molecule using single-particle cryo-EM. Moreover, the scattering potential from many hydrogen atoms can be visualized in difference maps, allowing a direct analysis of hydrogen-bonding networks. Our technological advances, combined with further approaches to accelerate data acquisition and improve sample quality, provide a route towards routine application of cryo-EM in high-throughput screening of small molecule modulators and structure-based drug discovery. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11657.map.gz emd_11657.map.gz | 167.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11657-v30.xml emd-11657-v30.xml emd-11657.xml emd-11657.xml | 23.5 KB 23.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11657_fsc.xml emd_11657_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_11657.png emd_11657.png | 71.8 KB | ||
| Masks |  emd_11657_msk_1.map emd_11657_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11657.cif.gz emd-11657.cif.gz | 7.2 KB | ||
| Others |  emd_11657_half_map_1.map.gz emd_11657_half_map_1.map.gz emd_11657_half_map_2.map.gz emd_11657_half_map_2.map.gz | 140.7 MB 140.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11657 http://ftp.pdbj.org/pub/emdb/structures/EMD-11657 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11657 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11657 | HTTPS FTP |
-Validation report
| Summary document |  emd_11657_validation.pdf.gz emd_11657_validation.pdf.gz | 826.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11657_full_validation.pdf.gz emd_11657_full_validation.pdf.gz | 826 KB | Display | |
| Data in XML |  emd_11657_validation.xml.gz emd_11657_validation.xml.gz | 20.2 KB | Display | |
| Data in CIF |  emd_11657_validation.cif.gz emd_11657_validation.cif.gz | 26.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11657 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11657 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11657 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11657 | HTTPS FTP |
-Related structure data
| Related structure data |  7a5vMC  7a4mC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10500 (Title: High resolution structure of GABAAR beta3 homopentamer EMPIAR-10500 (Title: High resolution structure of GABAAR beta3 homopentamerData size: 13.0 TB Data #1: CFEG, Falcon4, with 5 eV filter [micrographs - multiframe] Data #2: CFEG, Falcon4, with 5 eV filter, without objective aperture [micrographs - multiframe] Data #3: XFEG, Falcon3, without filter [micrographs - multiframe] Data #4: XFEG, Falcon4, with filter but retracted slit [micrographs - multiframe] Data #5: XFEG, Falcon4, with 3 eV filter [micrographs - multiframe] Data #6: XFEG, Falcon4, with 5 eV filter [micrographs - multiframe] Data #7: XFEG, Falcon4, with 3 eV filter [micrographs - multiframe] Data #8: XFEG, K3, without filter [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11657.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11657.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8117 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
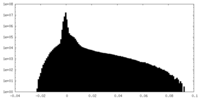
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11657_msk_1.map emd_11657_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
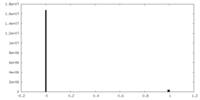
| Density Histograms |
-Half map: #2
| File | emd_11657_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
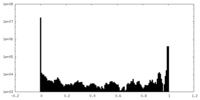
| Density Histograms |
-Half map: #1
| File | emd_11657_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
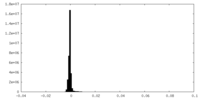
| Density Histograms |
- Sample components
Sample components
+Entire : Human GABA(A)R-beta3 homopentamer in complex with Megabody-25 in ...
+Supramolecule #1: Human GABA(A)R-beta3 homopentamer in complex with Megabody-25 in ...
+Supramolecule #2: Human GABA(A)R-beta3 homopentamer
+Supramolecule #3: Megabody-25
+Macromolecule #1: Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyri...
+Macromolecule #2: Megabody Mb25
+Macromolecule #5: HEXADECANE
+Macromolecule #6: DECANE
+Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
+Macromolecule #8: N-OCTANE
+Macromolecule #9: HISTAMINE
+Macromolecule #10: CHLORIDE ION
+Macromolecule #11: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 287 K / Instrument: LEICA PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.1 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 165000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7a5v: |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)