[English] 日本語
 Yorodumi
Yorodumi- EMDB-1160: Double hexameric ring assembly of the type III protein translocas... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1160 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
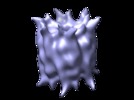
| Title | Double hexameric ring assembly of the type III protein translocase ATPase HrcN. | |||||||||
 Map data Map data | This volume contains the 3D reconstruction of dodecameric HrcN. It is D6-symmetrized. A threshold of 0.61 is recommended for surface rendering. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Pseudomonas syringae pv. phaseolicola (bacteria) Pseudomonas syringae pv. phaseolicola (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 16.0 Å | |||||||||
 Authors Authors | Muller SA / Poyidis C / Stone R / Meesters C / Chami M / Engel A / Economou A / Stahlberg H | |||||||||
 Citation Citation |  Journal: Mol Microbiol / Year: 2006 Journal: Mol Microbiol / Year: 2006Title: Double hexameric ring assembly of the type III protein translocase ATPase HrcN. Authors: Shirley A Müller / Charalambos Pozidis / Remington Stone / Christian Meesters / Mohamed Chami / Andreas Engel / Anastassios Economou / Henning Stahlberg /  Abstract: The specialized type III secretion (T3S) apparatus of pathogenic and symbiotic Gram-negative bacteria comprises a complex transmembrane organelle and an ATPase homologous to the F1-ATPase beta ...The specialized type III secretion (T3S) apparatus of pathogenic and symbiotic Gram-negative bacteria comprises a complex transmembrane organelle and an ATPase homologous to the F1-ATPase beta subunit. The T3S ATPase HrcN of Pseudomonas syringae associates with the inner membrane, and its ATP hydrolytic activity is stimulated by dodecamerization. The structure of dodecameric HrcN (HrcN12) determined to 1.6 nm by cryo-electron microscopy is presented. HrcN12 comprises two hexameric rings that are probably stacked face-to-face by the association of their C-terminal domains. It is 11.5 +/- 1.0 nm in diameter, 12.0 +/- 2.0 nm high and has a 2.0-3.8 nm wide inner channel. This structure is compared to a homology model based on the structure of the F1-beta-ATPase. A model for its incorporation within the T3S apparatus is presented. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1160.map.gz emd_1160.map.gz | 3.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1160-v30.xml emd-1160-v30.xml emd-1160.xml emd-1160.xml | 8.7 KB 8.7 KB | Display Display |  EMDB header EMDB header |
| Images |  1160.gif 1160.gif | 15.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1160 http://ftp.pdbj.org/pub/emdb/structures/EMD-1160 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1160 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1160 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1160.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1160.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This volume contains the 3D reconstruction of dodecameric HrcN. It is D6-symmetrized. A threshold of 0.61 is recommended for surface rendering. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : HrcN
| Entire | Name: HrcN |
|---|---|
| Components |
|
-Supramolecule #1000: HrcN
| Supramolecule | Name: HrcN / type: sample / ID: 1000 Details: The sample had a tendency to aggregate rapidly. Images were usually highly crowded, or empty, from the same cryo-EM grid. Oligomeric state: homododecamer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 601 KDa / Theoretical: 612 KDa / Method: STEM mass measurement |
-Macromolecule #1: HrcN
| Macromolecule | Name: HrcN / type: protein_or_peptide / ID: 1 / Name.synonym: HrcN / Number of copies: 12 / Oligomeric state: dodecamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Pseudomonas syringae pv. phaseolicola (bacteria) / synonym: Pseudomonas syringae pathovar phaseolicola Pseudomonas syringae pv. phaseolicola (bacteria) / synonym: Pseudomonas syringae pathovar phaseolicola |
| Molecular weight | Experimental: 612 KDa / Theoretical: 601 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 50mM Tris-Cl, pH 7.4, 0.2M NaCl, 20% glycerol |
| Grid | Details: Quantifoil 300 mesh |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: Home-built |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG |
|---|---|
| Image recording | Digitization - Scanner: PRIMESCAN / Digitization - Sampling interval: 5.0 µm / Number real images: 165 / Od range: 1 / Bits/pixel: 16 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 50000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 50000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: SPIDER |
|---|---|
| Final reconstruction | Applied symmetry - Point group: D6 (2x6 fold dihedral) / Resolution.type: BY AUTHOR / Resolution: 16.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER / Number images used: 8742 |
| Final two d classification | Number classes: 352 |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















