[English] 日本語
 Yorodumi
Yorodumi- EMDB-11437: Structure of the Paranosema locustae ribosome in complex with Lso2 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11437 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Paranosema locustae ribosome in complex with Lso2 | |||||||||
 Map data Map data | Combined focused maps, state 2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Microsporidia / Pathogen / Ribosome / Hibernation / Genome Compaction | |||||||||
| Biological species |  Paranosema locustae (fungus) Paranosema locustae (fungus) | |||||||||
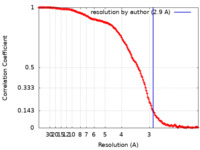
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Ehrenbolger K / Jespersen N / Sharma H / Sokolova YY / Tokarev YS / Vossbrinck CR / Barandun J | |||||||||
| Funding support |  Sweden, 1 items Sweden, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2020 Journal: PLoS Biol / Year: 2020Title: Differences in structure and hibernation mechanism highlight diversification of the microsporidian ribosome. Authors: Kai Ehrenbolger / Nathan Jespersen / Himanshu Sharma / Yuliya Y Sokolova / Yuri S Tokarev / Charles R Vossbrinck / Jonas Barandun /    Abstract: Assembling and powering ribosomes are energy-intensive processes requiring fine-tuned cellular control mechanisms. In organisms operating under strict nutrient limitations, such as pathogenic ...Assembling and powering ribosomes are energy-intensive processes requiring fine-tuned cellular control mechanisms. In organisms operating under strict nutrient limitations, such as pathogenic microsporidia, conservation of energy via ribosomal hibernation and recycling is critical. The mechanisms by which hibernation is achieved in microsporidia, however, remain poorly understood. Here, we present the cryo-electron microscopy structure of the ribosome from Paranosema locustae spores, bound by the conserved eukaryotic hibernation and recycling factor Lso2. The microsporidian Lso2 homolog adopts a V-shaped conformation to bridge the mRNA decoding site and the large subunit tRNA binding sites, providing a reversible ribosome inactivation mechanism. Although microsporidian ribosomes are highly compacted, the P. locustae ribosome retains several rRNA segments absent in other microsporidia, and represents an intermediate state of rRNA reduction. In one case, the near complete reduction of an expansion segment has resulted in a single bound nucleotide, which may act as an architectural co-factor to stabilize a protein-protein interface. The presented structure highlights the reductive evolution in these emerging pathogens and sheds light on a conserved mechanism for eukaryotic ribosome hibernation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11437.map.gz emd_11437.map.gz | 218 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11437-v30.xml emd-11437-v30.xml emd-11437.xml emd-11437.xml | 91.9 KB 91.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11437_fsc.xml emd_11437_fsc.xml | 18.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_11437.png emd_11437.png | 190 KB | ||
| Filedesc metadata |  emd-11437.cif.gz emd-11437.cif.gz | 17.5 KB | ||
| Others |  emd_11437_additional_1.map.gz emd_11437_additional_1.map.gz emd_11437_additional_2.map.gz emd_11437_additional_2.map.gz emd_11437_additional_3.map.gz emd_11437_additional_3.map.gz emd_11437_half_map_1.map.gz emd_11437_half_map_1.map.gz emd_11437_half_map_2.map.gz emd_11437_half_map_2.map.gz | 4.7 MB 16.7 MB 8.2 MB 217.8 MB 217.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11437 http://ftp.pdbj.org/pub/emdb/structures/EMD-11437 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11437 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11437 | HTTPS FTP |
-Related structure data
| Related structure data |  6zu5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-11076 (Title: Single particle cryo-EM dataset of the Paranosema locustae ribosome bound to Lso2 EMPIAR-11076 (Title: Single particle cryo-EM dataset of the Paranosema locustae ribosome bound to Lso2Data size: 2.0 TB Data #1: Unaligned multi-frame micrographs of the Paranosema locustae Lso2-ribosome complex - 20 frames [micrographs - multiframe] Data #2: Unaligned multi-frame micrographs of the Paranosema locustae Lso2-ribosome complex - 40 frames [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11437.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11437.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Combined focused maps, state 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Additional map 3 (SSU head focused)
| File | emd_11437_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional map 3 (SSU head focused) | ||||||||||||
| Projections & Slices |
| ||||||||||||
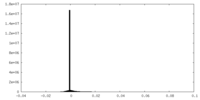
| Density Histograms |
-Additional map: Additional map 1 (LSU focused)
| File | emd_11437_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional map 1 (LSU focused) | ||||||||||||
| Projections & Slices |
| ||||||||||||
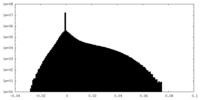
| Density Histograms |
-Additional map: Additional map 2 (SSU body focused)
| File | emd_11437_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Additional map 2 (SSU body focused) | ||||||||||||
| Projections & Slices |
| ||||||||||||
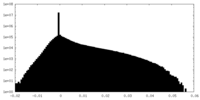
| Density Histograms |
-Half map: Combined focused half map 1, state 2
| File | emd_11437_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Combined focused half map 1, state 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
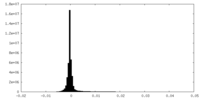
| Density Histograms |
-Half map: Combined focused half map 2, state 2
| File | emd_11437_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Combined focused half map 2, state 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Lso2 bound to the ribosome of Paranosema locustae
+Supramolecule #1: Lso2 bound to the ribosome of Paranosema locustae
+Macromolecule #1: 25S rRNA
+Macromolecule #2: 5S rRNA
+Macromolecule #43: 18S rRNA
+Macromolecule #3: uL2
+Macromolecule #4: uL15
+Macromolecule #5: uL3
+Macromolecule #6: eL29
+Macromolecule #7: uL4
+Macromolecule #8: eL30
+Macromolecule #9: uL18
+Macromolecule #10: eL31
+Macromolecule #11: eL6
+Macromolecule #12: eL32
+Macromolecule #13: uL30
+Macromolecule #14: eL33
+Macromolecule #15: eL8
+Macromolecule #16: eL34
+Macromolecule #17: uL6
+Macromolecule #18: uL29
+Macromolecule #19: uL16
+Macromolecule #20: eL36
+Macromolecule #21: uL5
+Macromolecule #22: eL37
+Macromolecule #23: eL13
+Macromolecule #24: eL39
+Macromolecule #25: eL14
+Macromolecule #26: eL40
+Macromolecule #27: eL15
+Macromolecule #28: Lso2
+Macromolecule #29: uL13
+Macromolecule #30: eL42
+Macromolecule #31: uL22
+Macromolecule #32: eL43
+Macromolecule #33: eL18
+Macromolecule #34: eL19
+Macromolecule #35: eL20
+Macromolecule #36: eL21
+Macromolecule #37: eL22
+Macromolecule #38: uL14
+Macromolecule #39: eL24
+Macromolecule #40: uL23
+Macromolecule #41: uL24
+Macromolecule #42: eL27
+Macromolecule #44: uS2
+Macromolecule #45: eS26
+Macromolecule #46: eS1
+Macromolecule #47: eS27
+Macromolecule #48: uS5
+Macromolecule #49: eS28
+Macromolecule #50: uS3
+Macromolecule #51: uS14
+Macromolecule #52: eS4
+Macromolecule #53: eS30
+Macromolecule #54: uS7
+Macromolecule #55: eS6
+Macromolecule #56: RACK1
+Macromolecule #57: eS7
+Macromolecule #58: eS8
+Macromolecule #59: uS4
+Macromolecule #60: eS10
+Macromolecule #61: uS17
+Macromolecule #62: uS15
+Macromolecule #63: uS11
+Macromolecule #64: uS19
+Macromolecule #65: uS9
+Macromolecule #66: eS17
+Macromolecule #67: uS13
+Macromolecule #68: eS19
+Macromolecule #69: uS10
+Macromolecule #70: eS21
+Macromolecule #71: uS8
+Macromolecule #72: uS12
+Macromolecule #73: eS24
+Macromolecule #74: eS25
+Macromolecule #75: MAGNESIUM ION
+Macromolecule #76: ZINC ION
+Macromolecule #77: ADENOSINE MONOPHOSPHATE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: 30 mM Tris-HCl, pH 7.5, 25 mM KCl, 5 mM magnesium acetate, 1 mM DTT, 1 mM EDTA |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Number grids imaged: 1 / Average electron dose: 28.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)