+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11376 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram averaging of the OSBP construct NPHFFAT. | |||||||||
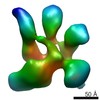
 Map data Map data | NPHFFAT in galactocerebroside tubes containing PI4P | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
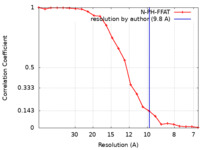
| Method | subtomogram averaging / cryo EM / Resolution: 9.8 Å | |||||||||
 Authors Authors | de la Mora E / Dezi M / diCicco A / Bigay J / Gautier R / Manzi J / Polidori J / Castano-Diez D / Mesmin B / Antonny B / Levy D | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Nanoscale architecture of a VAP-A-OSBP tethering complex at membrane contact sites. Authors: Eugenio de la Mora / Manuela Dezi / Aurélie Di Cicco / Joëlle Bigay / Romain Gautier / John Manzi / Joël Polidori / Daniel Castaño-Díez / Bruno Mesmin / Bruno Antonny / Daniel Lévy /   Abstract: Membrane contact sites (MCS) are subcellular regions where two organelles appose their membranes to exchange small molecules, including lipids. Structural information on how proteins form MCS is ...Membrane contact sites (MCS) are subcellular regions where two organelles appose their membranes to exchange small molecules, including lipids. Structural information on how proteins form MCS is scarce. We designed an in vitro MCS with two membranes and a pair of tethering proteins suitable for cryo-tomography analysis. It includes VAP-A, an ER transmembrane protein interacting with a myriad of cytosolic proteins, and oxysterol-binding protein (OSBP), a lipid transfer protein that transports cholesterol from the ER to the trans Golgi network. We show that VAP-A is a highly flexible protein, allowing formation of MCS of variable intermembrane distance. The tethering part of OSBP contains a central, dimeric, and helical T-shape region. We propose that the molecular flexibility of VAP-A enables the recruitment of partners of different sizes within MCS of adjustable thickness, whereas the T geometry of the OSBP dimer facilitates the movement of the two lipid-transfer domains between membranes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11376.map.gz emd_11376.map.gz | 504.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11376-v30.xml emd-11376-v30.xml emd-11376.xml emd-11376.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11376_fsc.xml emd_11376_fsc.xml | 2 KB | Display |  FSC data file FSC data file |
| Images |  emd_11376.png emd_11376.png | 42 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11376 http://ftp.pdbj.org/pub/emdb/structures/EMD-11376 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11376 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11376 | HTTPS FTP |
-Validation report
| Summary document |  emd_11376_validation.pdf.gz emd_11376_validation.pdf.gz | 343.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11376_full_validation.pdf.gz emd_11376_full_validation.pdf.gz | 342.8 KB | Display | |
| Data in XML |  emd_11376_validation.xml.gz emd_11376_validation.xml.gz | 6.2 KB | Display | |
| Data in CIF |  emd_11376_validation.cif.gz emd_11376_validation.cif.gz | 7.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11376 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11376 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11376 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11376 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_11376.map.gz / Format: CCP4 / Size: 549.8 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11376.map.gz / Format: CCP4 / Size: 549.8 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
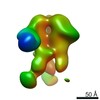
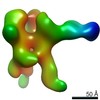
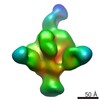
| Annotation | NPHFFAT in galactocerebroside tubes containing PI4P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Chimera N-PH-FFAT comprising residues 1-407 from the Oxysterol-Bi...
| Entire | Name: Chimera N-PH-FFAT comprising residues 1-407 from the Oxysterol-Binding Protein |
|---|---|
| Components |
|
-Supramolecule #1: Chimera N-PH-FFAT comprising residues 1-407 from the Oxysterol-Bi...
| Supramolecule | Name: Chimera N-PH-FFAT comprising residues 1-407 from the Oxysterol-Binding Protein type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 43.2 kDa/nm |
-Macromolecule #1: N-PH-FFAT (OSBP 1-407)
| Macromolecule | Name: N-PH-FFAT (OSBP 1-407) / type: other / ID: 1 Details: Chimera N-PH-FFAT comprising residues 1-407 from human Oxysterol-Binding Protein. Classification: other |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAATELRGVV GPGPAAIAAL GGGGAGPPVV GGGGGRGDAG PGSGAASGTV VAAAAGGPGP GAGGVAAAGP APAPPTGGSG GSGAGGSGSA REGWLFKWTN YIKGYQRRWF VLSNGLLSYY RSKAEMRHTC RGTINLATAN ITVEDSCNFI ISNGGAQTYH LKASSEVERQ ...String: MAATELRGVV GPGPAAIAAL GGGGAGPPVV GGGGGRGDAG PGSGAASGTV VAAAAGGPGP GAGGVAAAGP APAPPTGGSG GSGAGGSGSA REGWLFKWTN YIKGYQRRWF VLSNGLLSYY RSKAEMRHTC RGTINLATAN ITVEDSCNFI ISNGGAQTYH LKASSEVERQ RWVTALELAK AKAVKMLAES DESGDEESVS QTDKTELQNT LRTLSSKVED LSTCNDLIAK HGTALQRSLS ELESLKLPAE SNEKIKQVNE RATLFRITSN AMINACRDFL MLAQTHSKKW QKSLQYERDQ RIRLEETLEQ LAKQHNHLER AFRGATVLPA NTPGNVGSGK DQCCSGKGDM SDEDDENEFF DAPEIITMPE NLGHKRTGSN ISGASSDISL DEQYKHQLEE TKKEKRTR |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
Details: 50 mM HEPES pH 7.4 120 mM potassium acetate | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Homemade / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 98 % / Chamber temperature: 21 K / Instrument: LEICA PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 2 / Average electron dose: 3.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)