[English] 日本語
 Yorodumi
Yorodumi- EMDB-1135: Quaternary polymorphism of replicative helicase G40P: structural ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1135 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Quaternary polymorphism of replicative helicase G40P: structural mapping and domain rearrangement. | |||||||||
 Map data Map data | Bacteriophage SPP1 helicase G40P: C6 architecture | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Bacillus phage SPP1 (virus) Bacillus phage SPP1 (virus) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 24.0 Å | |||||||||
 Authors Authors | Nunez-Ramirez R / Robledo Y / mesa P / Alonso JC / Carazo JM / Donate LE | |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2006 Journal: J Mol Biol / Year: 2006Title: Quaternary polymorphism of replicative helicase G40P: structural mapping and domain rearrangement. Authors: Rafael Núñez-Ramírez / Yolanda Robledo / Pablo Mesa / Silvia Ayora / Juan Carlos Alonso / José María Carazo / Luis Enrique Donate /  Abstract: Quaternary polymorphism is a distinctive structural feature of the DnaB family of replicative DNA hexameric helicases. The Bacillus subtilis bacteriophage SPP1 gene 40 product (G40P) belongs to this ...Quaternary polymorphism is a distinctive structural feature of the DnaB family of replicative DNA hexameric helicases. The Bacillus subtilis bacteriophage SPP1 gene 40 product (G40P) belongs to this family. Three different quaternary states have been described for G40P homohexamers, two of them with C(3) symmetry, and the other with C(6) symmetry. We present three-dimensional reconstructions of the different architectures of G40P hexamers and a variant lacking the N-terminal domain. Comparison of the G40P and the deletion mutant structures sheds new light on the functional roles of the N and C-terminal domains, at the same time that it allows the direct structural mapping of these domains. Based on this new information, hybrid EM/X-ray models are presented for all the different symmetries. These results suggest that quaternary polymorphism of hexameric helicases may be implicated in the translocation along the DNA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1135.map.gz emd_1135.map.gz | 1.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1135-v30.xml emd-1135-v30.xml emd-1135.xml emd-1135.xml | 8.6 KB 8.6 KB | Display Display |  EMDB header EMDB header |
| Images |  1135.gif 1135.gif | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1135 http://ftp.pdbj.org/pub/emdb/structures/EMD-1135 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1135 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1135 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1135.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1135.map.gz / Format: CCP4 / Size: 1.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Bacteriophage SPP1 helicase G40P: C6 architecture | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
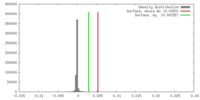
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : C6 architecture of Bacteriophage SPP1 replicative helicase G40P
| Entire | Name: C6 architecture of Bacteriophage SPP1 replicative helicase G40P |
|---|---|
| Components |
|
-Supramolecule #1000: C6 architecture of Bacteriophage SPP1 replicative helicase G40P
| Supramolecule | Name: C6 architecture of Bacteriophage SPP1 replicative helicase G40P type: sample / ID: 1000 / Oligomeric state: Homohexamer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 300 KDa / Theoretical: 300 KDa / Method: gel filtration chromatography |
-Macromolecule #1: helicase
| Macromolecule | Name: helicase / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Oligomeric state: Hexamer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Bacillus phage SPP1 (virus) / Strain: SPP1 / synonym: Bacteriphage SPP1 / Cell: Bacteria Bacillus phage SPP1 (virus) / Strain: SPP1 / synonym: Bacteriphage SPP1 / Cell: Bacteria |
| Molecular weight | Experimental: 300 KDa / Theoretical: 300 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7 Details: 50mM HEPES, 50mM NaCl, 10mM MgCl2, 1mM ATP, 1mM dithithreitol |
| Staining | Type: NEGATIVE Details: Grids with adsorbed protein satined on 2% uranyl acetate for 1 minute |
| Grid | Details: collodion/carbon coated 400 mesh copper grid |
| Vitrification | Cryogen name: NONE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2000EX |
|---|---|
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 21 µm / Number real images: 40 / Average electron dose: 20 e/Å2 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 80 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal magnification: 60000 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: OTHER |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C6 (6 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 24.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPIDER Xmipp / Number images used: 700 |
|---|
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera
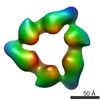
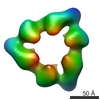
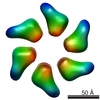


 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)