+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11159 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Dumbbell_1 | |||||||||
 Map data Map data | dumbbell-shaped DNA origami object | |||||||||
 Sample Sample |
| |||||||||
| Biological species | synthetic construct (others) | |||||||||
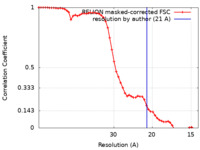
| Method | single particle reconstruction / cryo EM / Resolution: 21.0 Å | |||||||||
 Authors Authors | Kube M / Kohler F / Feigl E / Nagel-Yuksel B / Willner EM / Funke JJ / Gerling T / Stommer P / Honemann MN / Martin TG ...Kube M / Kohler F / Feigl E / Nagel-Yuksel B / Willner EM / Funke JJ / Gerling T / Stommer P / Honemann MN / Martin TG / Scheres SHW / Dietz H | |||||||||
| Funding support |  Germany, European Union, 2 items Germany, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution. Authors: Massimo Kube / Fabian Kohler / Elija Feigl / Baki Nagel-Yüksel / Elena M Willner / Jonas J Funke / Thomas Gerling / Pierre Stömmer / Maximilian N Honemann / Thomas G Martin / Sjors H W ...Authors: Massimo Kube / Fabian Kohler / Elija Feigl / Baki Nagel-Yüksel / Elena M Willner / Jonas J Funke / Thomas Gerling / Pierre Stömmer / Maximilian N Honemann / Thomas G Martin / Sjors H W Scheres / Hendrik Dietz /   Abstract: The methods of DNA nanotechnology enable the rational design of custom shapes that self-assemble in solution from sets of DNA molecules. DNA origami, in which a long template DNA single strand is ...The methods of DNA nanotechnology enable the rational design of custom shapes that self-assemble in solution from sets of DNA molecules. DNA origami, in which a long template DNA single strand is folded by many short DNA oligonucleotides, can be employed to make objects comprising hundreds of unique DNA strands and thousands of base pairs, thus in principle providing many degrees of freedom for modelling complex objects of defined 3D shapes and sizes. Here, we address the problem of accurate structural validation of DNA objects in solution with cryo-EM based methodologies. By taking into account structural fluctuations, we can determine structures with improved detail compared to previous work. To interpret the experimental cryo-EM maps, we present molecular-dynamics-based methods for building pseudo-atomic models in a semi-automated fashion. Among other features, our data allows discerning details such as helical grooves, single-strand versus double-strand crossovers, backbone phosphate positions, and single-strand breaks. Obtaining this higher level of detail is a step forward that now allows designers to inspect and refine their designs with base-pair level interventions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11159.map.gz emd_11159.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11159-v30.xml emd-11159-v30.xml emd-11159.xml emd-11159.xml | 12 KB 12 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11159_fsc.xml emd_11159_fsc.xml | 5.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_11159.png emd_11159.png | 47.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11159 http://ftp.pdbj.org/pub/emdb/structures/EMD-11159 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11159 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11159 | HTTPS FTP |
-Related structure data
| Related structure data |  7areC  7arqC  7artC  7arvC  7aryC  7as5C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11159.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11159.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | dumbbell-shaped DNA origami object | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 7.16 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : dumbbell-shaped multilayer DNA nanostructure without crossovers a...
| Entire | Name: dumbbell-shaped multilayer DNA nanostructure without crossovers at the "handle" |
|---|---|
| Components |
|
-Supramolecule #1: dumbbell-shaped multilayer DNA nanostructure without crossovers a...
| Supramolecule | Name: dumbbell-shaped multilayer DNA nanostructure without crossovers at the "handle" type: complex / ID: 1 / Parent: 0 / Details: DNA Origami |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Recombinant expression | Organism: synthetic construct (others) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
| Details | monodisperse, 1300nM |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: CEOS Cs corrector |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number real images: 220 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 4.717 µm / Calibrated defocus min: 1.192 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal magnification: 37000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)