[English] 日本語
 Yorodumi
Yorodumi- EMDB-1107: Structural basis of pore formation by the bacterial toxin pneumolysin. -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1107 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural basis of pore formation by the bacterial toxin pneumolysin. | |||||||||
 Map data Map data | This is the 3D map for the pore form of pneumolysin, calculated using c38 symmetry. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationhemolysis in another organism / cholesterol binding / membrane => GO:0016020 / toxin activity / host cell plasma membrane / extracellular region / membrane Similarity search - Function | |||||||||
| Biological species | synthetic construct (others) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 29.0 Å | |||||||||
 Authors Authors | Tilley SJ / Orlova EV / Gilbert RJ / Andrew PW / Saibil HR | |||||||||
 Citation Citation |  Journal: Cell / Year: 2005 Journal: Cell / Year: 2005Title: Structural basis of pore formation by the bacterial toxin pneumolysin. Authors: Sarah J Tilley / Elena V Orlova / Robert J C Gilbert / Peter W Andrew / Helen R Saibil /  Abstract: The bacterial toxin pneumolysin is released as a soluble monomer that kills target cells by assembling into large oligomeric rings and forming pores in cholesterol-containing membranes. Using cryo-EM ...The bacterial toxin pneumolysin is released as a soluble monomer that kills target cells by assembling into large oligomeric rings and forming pores in cholesterol-containing membranes. Using cryo-EM and image processing, we have determined the structures of membrane-surface bound (prepore) and inserted-pore oligomer forms, providing a direct observation of the conformational transition into the pore form of a cholesterol-dependent cytolysin. In the pore structure, the domains of the monomer separate and double over into an arch, forming a wall sealing the bilayer around the pore. This transformation is accomplished by substantial refolding of two of the four protein domains along with deformation of the membrane. Extension of protein density into the bilayer supports earlier predictions that the protein inserts beta hairpins into the membrane. With an oligomer size of up to 44 subunits in the pore, this assembly creates a transmembrane channel 260 A in diameter lined by 176 beta strands. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1107.map.gz emd_1107.map.gz | 1.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1107-v30.xml emd-1107-v30.xml emd-1107.xml emd-1107.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| Images |  1107.gif 1107.gif | 62.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1107 http://ftp.pdbj.org/pub/emdb/structures/EMD-1107 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1107 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1107 | HTTPS FTP |
-Related structure data
| Related structure data |  2bk1MC  1106C  1108C  2bk2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1107.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1107.map.gz / Format: CCP4 / Size: 15.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the 3D map for the pore form of pneumolysin, calculated using c38 symmetry. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.5 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
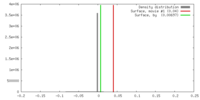
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Pneumolysin
| Entire | Name: Pneumolysin |
|---|---|
| Components |
|
-Supramolecule #1000: Pneumolysin
| Supramolecule | Name: Pneumolysin / type: sample / ID: 1000 / Oligomeric state: 38-mer / Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 2.0 MDa |
-Supramolecule #1: Phosphatidylcholine-cholesterol lipid bilayer
| Supramolecule | Name: Phosphatidylcholine-cholesterol lipid bilayer / type: organelle_or_cellular_component / ID: 1 / Recombinant expression: No |
|---|---|
| Source (natural) | Organism: synthetic construct (others) / synonym: membrane |
-Macromolecule #1: Pneumolysin
| Macromolecule | Name: Pneumolysin / type: protein_or_peptide / ID: 1 / Number of copies: 38 / Oligomeric state: 38-mer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 2.0 MDa |
| Recombinant expression | Organism:  |
| Sequence | InterPro: Thiol-activated cytolysin |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 6.95 Details: 8 mM Na2HPO4, 1.5 mM KH2PO4, 2.5 mM KCl, 0.25M NaCl |
| Grid | Details: holey carbon 400 mesh copper grid, glow discharged using positive charge |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 96 % / Chamber temperature: 100 K / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: home made plunger Method: Grids were blotted for approximately 3 seconds and allowed to drain vertically for 5 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Temperature | Min: 100 K / Max: 100 K / Average: 100 K |
| Alignment procedure | Legacy - Astigmatism: corrected at 150,000 magnification |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 135 / Average electron dose: 20 e/Å2 / Details: After scanning images were averaged 2x2. / Od range: 1 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.1 µm / Nominal magnification: 42000 |
| Sample stage | Specimen holder: Side entry / Specimen holder model: GATAN LIQUID NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Details | The 1pfo structure was separated into six rigid bodies: domain 1 (91-172, 231-272, 354-373), domain 2 upper (53-62, 83-90, 374-381), domain 2 lower (63-82, 382-390), domain 3 (177-186, 221-230, 273-283, 316-353), domain 3 hairpins (187-220, 284-315), and domain 4 (391-500). These rigid bodies were fitted manually using the software O and pymol. |
| Refinement | Protocol: RIGID BODY FIT |
| Output model |  PDB-2bk1: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)