[English] 日本語
 Yorodumi
Yorodumi- EMDB-11047: In situ cryo-electron tomography reveals layered organization of ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11047 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | In situ cryo-electron tomography reveals layered organization of pre-ribosome maturation in nucleoli. Large Subunit Pre-Ribosome, Class 2 | |||||||||
 Map data Map data | Chlamydomonas reinhardtii Large Subunit Pre-Ribosome, Class 2 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Erdmann PS / Klumpe S / Hou Z / Beck F / Wilfling F / Plitzko JM / Baumeister W | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: In situ cryo-electron tomography reveals gradient organization of ribosome biogenesis in intact nucleoli. Authors: Philipp S Erdmann / Zhen Hou / Sven Klumpe / Sagar Khavnekar / Florian Beck / Florian Wilfling / Jürgen M Plitzko / Wolfgang Baumeister /   Abstract: Ribosomes comprise a large (LSU) and a small subunit (SSU) which are synthesized independently in the nucleolus before being exported into the cytoplasm, where they assemble into functional ribosomes. ...Ribosomes comprise a large (LSU) and a small subunit (SSU) which are synthesized independently in the nucleolus before being exported into the cytoplasm, where they assemble into functional ribosomes. Individual maturation steps have been analyzed in detail using biochemical methods, light microscopy and conventional electron microscopy (EM). In recent years, single particle analysis (SPA) has yielded molecular resolution structures of several pre-ribosomal intermediates. It falls short, however, of revealing the spatiotemporal sequence of ribosome biogenesis in the cellular context. Here, we present our study on native nucleoli in Chlamydomonas reinhardtii, in which we follow the formation of LSU and SSU precursors by in situ cryo-electron tomography (cryo-ET) and subtomogram averaging (STA). By combining both positional and molecular data, we reveal gradients of ribosome maturation within the granular component (GC), offering a new perspective on how the liquid-liquid-phase separation of the nucleolus supports ribosome biogenesis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11047.map.gz emd_11047.map.gz | 28.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11047-v30.xml emd-11047-v30.xml emd-11047.xml emd-11047.xml | 9.7 KB 9.7 KB | Display Display |  EMDB header EMDB header |
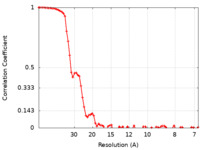
| FSC (resolution estimation) |  emd_11047_fsc.xml emd_11047_fsc.xml | 7.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_11047.png emd_11047.png | 57.1 KB | ||
| Masks |  emd_11047_msk_1.map emd_11047_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11047 http://ftp.pdbj.org/pub/emdb/structures/EMD-11047 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11047 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11047 | HTTPS FTP |
-Validation report
| Summary document |  emd_11047_validation.pdf.gz emd_11047_validation.pdf.gz | 245.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11047_full_validation.pdf.gz emd_11047_full_validation.pdf.gz | 244.9 KB | Display | |
| Data in XML |  emd_11047_validation.xml.gz emd_11047_validation.xml.gz | 9.8 KB | Display | |
| Data in CIF |  emd_11047_validation.cif.gz emd_11047_validation.cif.gz | 12.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11047 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11047 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11047 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11047 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11047.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11047.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Chlamydomonas reinhardtii Large Subunit Pre-Ribosome, Class 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.42 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11047_msk_1.map emd_11047_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : nucleolar small subunit processome, class 1
| Entire | Name: nucleolar small subunit processome, class 1 |
|---|---|
| Components |
|
-Supramolecule #1: nucleolar small subunit processome, class 1
| Supramolecule | Name: nucleolar small subunit processome, class 1 / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
| Details | in situ focus ion beam milled cells |
| Sectioning | Other: NO SECTIONING |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.5 e/Å2 Details: mixture of dose-symmetric, and bidirectional tilting schemes |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)