[English] 日本語
 Yorodumi
Yorodumi- EMDB-1097: Nuclear pore complex structure and dynamics revealed by cryoelect... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1097 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. | |||||||||
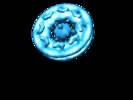
 Map data Map data | 3D structure of the Dictyostelium nuclear pore complex obtained by Cryoelectron Tomography | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 85.0 Å | |||||||||
 Authors Authors | Beck M / Forster F / Ecke M / Plitzko JM / Melchior F / Gerisch G / Baumeister W / Medalia O | |||||||||
 Citation Citation |  Journal: Science / Year: 2004 Journal: Science / Year: 2004Title: Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Authors: Martin Beck / Friedrich Förster / Mary Ecke / Jürgen M Plitzko / Frauke Melchior / Günther Gerisch / Wolfgang Baumeister / Ohad Medalia /  Abstract: Nuclear pore complexes (NPCs) are gateways for nucleocytoplasmic exchange. To analyze their structure in a close-to-life state, we studied transport-active, intact nuclei from Dictyostelium ...Nuclear pore complexes (NPCs) are gateways for nucleocytoplasmic exchange. To analyze their structure in a close-to-life state, we studied transport-active, intact nuclei from Dictyostelium discoideum by means of cryoelectron tomography. Subvolumes of the tomograms containing individual NPCs were extracted in silico and subjected to three-dimensional classification and averaging, whereby distinct structural states were observed. The central plug/transporter (CP/T) was variable in volume and could occupy different positions along the nucleocytoplasmic axis, which supports the notion that it essentially represents cargo in transit. Changes in the position of the CP/T were accompanied by structural rearrangements in the NPC scaffold. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1097.map.gz emd_1097.map.gz | 1.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1097-v30.xml emd-1097-v30.xml emd-1097.xml emd-1097.xml | 8.5 KB 8.5 KB | Display Display |  EMDB header EMDB header |
| Images |  1097.gif 1097.gif | 49 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1097 http://ftp.pdbj.org/pub/emdb/structures/EMD-1097 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1097 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1097 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1097.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1097.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D structure of the Dictyostelium nuclear pore complex obtained by Cryoelectron Tomography | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 16.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : nuclear pore complex
| Entire | Name: nuclear pore complex |
|---|---|
| Components |
|
-Supramolecule #1000: nuclear pore complex
| Supramolecule | Name: nuclear pore complex / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Macromolecule #1: nuclear pore complex
| Macromolecule | Name: nuclear pore complex / type: protein_or_peptide / ID: 1 / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | Details: 50 mM Tris pH 7.6, 25 mM KCl, 5mM MgCl2, 50 mM sucrose |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER / Details: Vitrification instrument: Rudolf Gatz plunger |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM300FEG/T |
|---|---|
| Temperature | Average: 90 K |
| Specialist optics | Energy filter - Name: GIF2000 / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 20.0 eV |
| Image recording | Category: CCD / Film or detector model: GATAN MULTISCAN |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal magnification: 17500 |
| Sample stage | Specimen holder: Eucentric / Specimen holder model: GATAN LIQUID NITROGEN / Tilt series - Axis1 - Max angle: 63 ° |
- Image processing
Image processing
| Details | Average number of projections used in the 3D reconstructions: 267. |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 85.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: TOM / Details: 3D reconstruction by averaging of subtomograms |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















