[English] 日本語
 Yorodumi
Yorodumi- EMDB-10925: Cryo-EM structure of undecameric human CALHM6 in the presence of Ca2+ -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10925 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of undecameric human CALHM6 in the presence of Ca2+ | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | calcium homeostasis modulator / ion channel / placenta / MEMBRANE PROTEIN | |||||||||
| Function / homology | regulation of natural killer cell cytokine production / voltage-gated channel activity / Calcium homeostasis modulator family / Calcium homeostasis modulator / ATP export / immunological synapse / monoatomic cation channel activity / plasma membrane / Calcium homeostasis modulator protein 6 Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
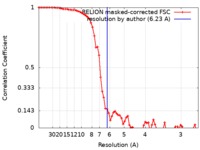
| Method | single particle reconstruction / cryo EM / Resolution: 6.23 Å | |||||||||
 Authors Authors | Sawicka M / Drozdzyk K | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Cryo-EM structures and functional properties of CALHM channels of the human placenta. Authors: Katarzyna Drożdżyk / Marta Sawicka / Maria-Isabel Bahamonde-Santos / Zaugg Jonas / Dawid Deneka / Christiane Albrecht / Raimund Dutzler /  Abstract: The transport of substances across the placenta is essential for the development of the fetus. Here, we were interested in the role of channels of the calcium homeostasis modulator (CALHM) family in ...The transport of substances across the placenta is essential for the development of the fetus. Here, we were interested in the role of channels of the calcium homeostasis modulator (CALHM) family in the human placenta. By transcript analysis, we found the paralogs CALHM2, 4, and 6 to be highly expressed in this organ and upregulated during trophoblast differentiation. Based on electrophysiology, we observed that activation of these paralogs differs from the voltage- and calcium-gated channel CALHM1. Cryo-EM structures of CALHM4 display decameric and undecameric assemblies with large cylindrical pore, while in CALHM6 a conformational change has converted the pore shape into a conus that narrows at the intracellular side, thus describing distinct functional states of the channel. The pore geometry alters the distribution of lipids, which occupy the cylindrical pore of CALHM4 in a bilayer-like arrangement whereas they have redistributed in the conical pore of CALHM6 with potential functional consequences. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10925.map.gz emd_10925.map.gz | 4.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10925-v30.xml emd-10925-v30.xml emd-10925.xml emd-10925.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10925_fsc.xml emd_10925_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_10925.png emd_10925.png | 90.8 KB | ||
| Masks |  emd_10925_msk_1.map emd_10925_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10925.cif.gz emd-10925.cif.gz | 5.8 KB | ||
| Others |  emd_10925_half_map_1.map.gz emd_10925_half_map_1.map.gz emd_10925_half_map_2.map.gz emd_10925_half_map_2.map.gz | 23.3 MB 23.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10925 http://ftp.pdbj.org/pub/emdb/structures/EMD-10925 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10925 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10925 | HTTPS FTP |
-Validation report
| Summary document |  emd_10925_validation.pdf.gz emd_10925_validation.pdf.gz | 668 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10925_full_validation.pdf.gz emd_10925_full_validation.pdf.gz | 667.5 KB | Display | |
| Data in XML |  emd_10925_validation.xml.gz emd_10925_validation.xml.gz | 12.9 KB | Display | |
| Data in CIF |  emd_10925_validation.cif.gz emd_10925_validation.cif.gz | 17.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10925 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10925 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10925 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10925 | HTTPS FTP |
-Related structure data
| Related structure data |  6ytxMC  6ytkC  6ytlC  6ytoC  6ytqC  6ytvC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10925.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10925.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10925_msk_1.map emd_10925_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_10925_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_10925_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Undecameric CALHM6
| Entire | Name: Undecameric CALHM6 |
|---|---|
| Components |
|
-Supramolecule #1: Undecameric CALHM6
| Supramolecule | Name: Undecameric CALHM6 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Calcium homeostasis modulator protein 6
| Macromolecule | Name: Calcium homeostasis modulator protein 6 / type: protein_or_peptide / ID: 1 / Number of copies: 11 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 34.494109 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEKFRAVLDL HVKHHSALGY GLVTLLTAGG ERIFSAVAFQ CPCSAAWNLP YGLVFLLVPA LALFLLGYVL SARTWRLLTG CCSSARASC GSALRGSLVC TQISAAAALA PLTWVAVALL GGAFYECAAT GSAAFAQRLC LGRNRSCAAE LPLVPCNQAK A SDVQDLLK ...String: MEKFRAVLDL HVKHHSALGY GLVTLLTAGG ERIFSAVAFQ CPCSAAWNLP YGLVFLLVPA LALFLLGYVL SARTWRLLTG CCSSARASC GSALRGSLVC TQISAAAALA PLTWVAVALL GGAFYECAAT GSAAFAQRLC LGRNRSCAAE LPLVPCNQAK A SDVQDLLK DLKAQSQVLG WILIAVVIII LLIFTSVTRC LSPVSFLQLK FWKIYLEQEQ QILKSKATEH ATELAKENIK CF FEGSHPK EYNTPSMKEW QQISSLYTFN PKGQYYSMLH KYVNRKEKTH SIRSTEGDTV IPVLGFVDSS GINSTPEL UniProtKB: Calcium homeostasis modulator protein 6 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)