+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10903 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human TRPC5 in complex with Pico145 (HC-608) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ion channel / small molecule / inhibitor / tetramer / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of membrane hyperpolarization / phosphatidylserine exposure on apoptotic cell surface / negative regulation of dendrite morphogenesis / Role of second messengers in netrin-1 signaling / store-operated calcium channel activity / inositol 1,4,5 trisphosphate binding / cation channel complex / actinin binding / TRP channels / clathrin binding ...regulation of membrane hyperpolarization / phosphatidylserine exposure on apoptotic cell surface / negative regulation of dendrite morphogenesis / Role of second messengers in netrin-1 signaling / store-operated calcium channel activity / inositol 1,4,5 trisphosphate binding / cation channel complex / actinin binding / TRP channels / clathrin binding / detection of maltose stimulus / maltose transport complex / carbohydrate transport / carbohydrate transmembrane transporter activity / maltose binding / maltose transport / maltodextrin transmembrane transport / regulation of cytosolic calcium ion concentration / positive regulation of axon extension / ATP-binding cassette (ABC) transporter complex, substrate-binding subunit-containing / calcium channel complex / positive regulation of neuron differentiation / ATP-binding cassette (ABC) transporter complex / cell chemotaxis / calcium ion transmembrane transport / calcium channel activity / neuron differentiation / calcium ion transport / nervous system development / presynapse / actin binding / outer membrane-bounded periplasmic space / growth cone / positive regulation of cytosolic calcium ion concentration / ATPase binding / neuron apoptotic process / periplasmic space / neuronal cell body / positive regulation of cell population proliferation / dendrite / DNA damage response / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
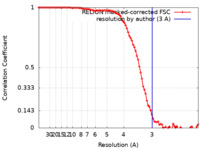
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Wright DJ / Johnson RM | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2020 Journal: Commun Biol / Year: 2020Title: Human TRPC5 structures reveal interaction of a xanthine-based TRPC1/4/5 inhibitor with a conserved lipid binding site. Authors: David J Wright / Katie J Simmons / Rachel M Johnson / David J Beech / Stephen P Muench / Robin S Bon /  Abstract: TRPC1/4/5 channels are non-specific cation channels implicated in a wide variety of diseases, and TRPC1/4/5 inhibitors have recently entered clinical trials. However, fundamental and translational ...TRPC1/4/5 channels are non-specific cation channels implicated in a wide variety of diseases, and TRPC1/4/5 inhibitors have recently entered clinical trials. However, fundamental and translational studies require a better understanding of TRPC1/4/5 channel regulation by endogenous and exogenous factors. Although several potent and selective TRPC1/4/5 modulators have been reported, the paucity of mechanistic insights into their modes-of-action remains a barrier to the development of new chemical probes and drug candidates. Xanthine-based modulators include the most potent and selective TRPC1/4/5 inhibitors described to date, as well as TRPC5 activators. Our previous studies suggest that xanthines interact with a, so far, elusive pocket of TRPC1/4/5 channels that is essential to channel gating. Here we report the structure of a small-molecule-bound TRPC1/4/5 channel-human TRPC5 in complex with the xanthine Pico145-to 3.0 Å. We found that Pico145 binds to a conserved lipid binding site of TRPC5, where it displaces a bound phospholipid. Our findings explain the mode-of-action of xanthine-based TRPC1/4/5 modulators, and suggest a structural basis for TRPC1/4/5 modulation by endogenous factors such as (phospho)lipids and Zn ions. These studies lay the foundations for the structure-based design of new generations of TRPC1/4/5 modulators. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10903.map.gz emd_10903.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10903-v30.xml emd-10903-v30.xml emd-10903.xml emd-10903.xml | 12.8 KB 12.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10903_fsc.xml emd_10903_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_10903.png emd_10903.png | 80.6 KB | ||
| Filedesc metadata |  emd-10903.cif.gz emd-10903.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10903 http://ftp.pdbj.org/pub/emdb/structures/EMD-10903 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10903 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10903 | HTTPS FTP |
-Validation report
| Summary document |  emd_10903_validation.pdf.gz emd_10903_validation.pdf.gz | 582.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10903_full_validation.pdf.gz emd_10903_full_validation.pdf.gz | 582.2 KB | Display | |
| Data in XML |  emd_10903_validation.xml.gz emd_10903_validation.xml.gz | 10.8 KB | Display | |
| Data in CIF |  emd_10903_validation.cif.gz emd_10903_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10903 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10903 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10903 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10903 | HTTPS FTP |
-Related structure data
| Related structure data |  6ysnMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10903.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10903.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Homotetrameric TRPC5
| Entire | Name: Homotetrameric TRPC5 |
|---|---|
| Components |
|
-Supramolecule #1: Homotetrameric TRPC5
| Supramolecule | Name: Homotetrameric TRPC5 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Maltose/maltodextrin-binding periplasmic protein,Short transient ...
| Macromolecule | Name: Maltose/maltodextrin-binding periplasmic protein,Short transient receptor potential channel 5 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 130.760031 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKIEEGKLVI WINGDKGYNG LAEVGKKFEK DTGIKVTVEH PDKLEEKFPQ VAATGDGPDI IFWAHDRFGG YAQSGLLAEI TPDKAFQDK LYPFTWDAVR YNGKLIAYPI AVEALSLIYN KDLLPNPPKT WEEIPALDKE LKAKGKSALM FNLQEPYFTW P LIAADGGY ...String: MKIEEGKLVI WINGDKGYNG LAEVGKKFEK DTGIKVTVEH PDKLEEKFPQ VAATGDGPDI IFWAHDRFGG YAQSGLLAEI TPDKAFQDK LYPFTWDAVR YNGKLIAYPI AVEALSLIYN KDLLPNPPKT WEEIPALDKE LKAKGKSALM FNLQEPYFTW P LIAADGGY AFKYENGKYD IKDVGVDNAG AKAGLTFLVD LIKNKHMNAD TDYSIAEAAF NKGETAMTIN GPWAWSNIDT SK VNYGVTV LPTFKGQPSK PFVGVLSAGI NAASPNKELA KEFLENYLLT DEGLEAVNKD KPLGAVALKS YEEELAKDPR IAA TMENAQ KGEIMPNIPQ MSAFWYAVRT AVINAASGLE VRQTVDEALK DAQTSSGLGV LFQGPMAQLY YKKVNYSPYR DRIP LQIVR AETELSAEEK AFLNAVEKGD YATVKQALQE AEIYYNVNIN CMDPLGRSAL LIAIENENLE IMELLLNHSV YVGDA LLYA IRKEVVGAVE LLLSYRRPSG EKQVPTLMMD TQFSEFTPDI TPIMLAAHTN NYEIIKLLVQ KRVTIPRPHQ IRCNCV ECV SSSEVDSLRH SRSRLNIYKA LASPSLIALS SEDPILTAFR LGWELKELSK VENEFKAEYE ELSQQCKLFA KDLLDQA RS SRELEIILNH RDDHSEELDP QKYHDLAKLK VAIKYHQKEF VAQPNCQQLL ATLWYDGFPG WRRKHWVVKL LTCMTIGF L FPMLSIAYLI SPRSNLGLFI KKPFIKFICH TASYLTFLFM LLLASQHIVR TDLHVQGPPP TVVEWMILPW VLGFIWGEI KEMWDGGFTE YIHDWWNLMD FAMNSLYLAT ISLKIMAYVK YNGSRPREEW EMWHPTLIAE ALFAISNILS SLRLISLFTA NSHLGPLQI SLGRMLLDIL KFLFIYCLVL LAFANGLNQL YFYYETRAID EPNNCKGIRC EKQNNAFSTL FETLQSLFWS V FGLLNLYV TNVKARHEFT EFVGATMFGT YNVISLVVLL NMLIAMMNNS YQLIADHADI EWKFARTKLW MSYFDEGGTL PP PFNIIPS PKSFLYLGNW FNNTFCPKRD PDGRRRRRNL RSFTERNADS LIQNQHYQEV IRNLVKRYVA AMIRNSKTHE GLT EENFKE LKQDISSFRY EVLDLLGNRK UniProtKB: Maltose/maltodextrin-binding periplasmic protein, Short transient receptor potential channel 5 |
-Macromolecule #2: 7-[(4-chlorophenyl)methyl]-3-methyl-1-(3-oxidanylpropyl)-8-[3-(tr...
| Macromolecule | Name: 7-[(4-chlorophenyl)methyl]-3-methyl-1-(3-oxidanylpropyl)-8-[3-(trifluoromethyloxy)phenoxy]purine-2,6-dione type: ligand / ID: 2 / Number of copies: 4 / Formula: PJQ |
|---|---|
| Molecular weight | Theoretical: 524.877 Da |
| Chemical component information |  ChemComp-PJQ: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE | |||||||||||||||
| Details | Final sample in PMAL-C8 amphipol |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 10.0 sec. / Average electron dose: 75.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: -0.003 µm / Nominal defocus min: -0.001 µm / Nominal magnification: 130000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)