[English] 日本語
 Yorodumi
Yorodumi- EMDB-10833: Negative stain reconstruction of grafix crosslink human condensin... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10833 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain reconstruction of grafix crosslink human condensin II in the presence of ATPyS | |||||||||
 Map data Map data | Human condensin II negative stain reconstruction, crosslinked with glutaraldehyde in the presence of ATPyS in a glycerol gradient | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
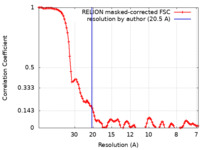
| Method | single particle reconstruction / negative staining / Resolution: 20.5 Å | |||||||||
 Authors Authors | Cutts EE / Beuron F / Morris E / Vannini A | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2020 Journal: Mol Cell / Year: 2020Title: Human Condensin I and II Drive Extensive ATP-Dependent Compaction of Nucleosome-Bound DNA. Authors: Muwen Kong / Erin E Cutts / Dongqing Pan / Fabienne Beuron / Thangavelu Kaliyappan / Chaoyou Xue / Edward P Morris / Andrea Musacchio / Alessandro Vannini / Eric C Greene /     Abstract: Structural maintenance of chromosomes (SMC) complexes are essential for genome organization from bacteria to humans, but their mechanisms of action remain poorly understood. Here, we characterize ...Structural maintenance of chromosomes (SMC) complexes are essential for genome organization from bacteria to humans, but their mechanisms of action remain poorly understood. Here, we characterize human SMC complexes condensin I and II and unveil the architecture of the human condensin II complex, revealing two putative DNA-entrapment sites. Using single-molecule imaging, we demonstrate that both condensin I and II exhibit ATP-dependent motor activity and promote extensive and reversible compaction of double-stranded DNA. Nucleosomes are incorporated into DNA loops during compaction without being displaced from the DNA, indicating that condensin complexes can readily act upon nucleosome-bound DNA molecules. These observations shed light on critical processes involved in genome organization in human cells. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10833.map.gz emd_10833.map.gz | 221 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10833-v30.xml emd-10833-v30.xml emd-10833.xml emd-10833.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10833_fsc.xml emd_10833_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_10833.png emd_10833.png | 23.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10833 http://ftp.pdbj.org/pub/emdb/structures/EMD-10833 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10833 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10833 | HTTPS FTP |
-Validation report
| Summary document |  emd_10833_validation.pdf.gz emd_10833_validation.pdf.gz | 203.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10833_full_validation.pdf.gz emd_10833_full_validation.pdf.gz | 203 KB | Display | |
| Data in XML |  emd_10833_validation.xml.gz emd_10833_validation.xml.gz | 10.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10833 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10833 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10833 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10833 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10833.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10833.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human condensin II negative stain reconstruction, crosslinked with glutaraldehyde in the presence of ATPyS in a glycerol gradient | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.19 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human condensin II pentamer composed of SMC2, SMC4, CapH2, CapD3 ...
| Entire | Name: Human condensin II pentamer composed of SMC2, SMC4, CapH2, CapD3 and CapG2 |
|---|---|
| Components |
|
-Supramolecule #1: Human condensin II pentamer composed of SMC2, SMC4, CapH2, CapD3 ...
| Supramolecule | Name: Human condensin II pentamer composed of SMC2, SMC4, CapH2, CapD3 and CapG2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Molecular weight | Theoretical: 657 KDa |
-Macromolecule #1: SMC2
| Macromolecule | Name: SMC2 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHIKSIILEG FKSYAQRTEV NGFDPLFNA ITGLNGSGKS N ILDSICFL LGISNLSQVR AS NLQDLVY KNGQAGITKA SVS ITFDNS DKKQSPLGFE VHDE ITVTR QVVIGGRNKY LINGV NANN TRVQDLFCSV GLNVNN PHF LIMQGRITKV LNMKPPE IL ...String: MHIKSIILEG FKSYAQRTEV NGFDPLFNA ITGLNGSGKS N ILDSICFL LGISNLSQVR AS NLQDLVY KNGQAGITKA SVS ITFDNS DKKQSPLGFE VHDE ITVTR QVVIGGRNKY LINGV NANN TRVQDLFCSV GLNVNN PHF LIMQGRITKV LNMKPPE IL SMIEEAAGTR MYEYKKIA A QKTIEKKEAK LKEIKTILE EEITPTIQKL KEERSSYLEY QKVMREIEH LSRLYIAYQF L LAEDTKVR SAEELKEMQD KV IKLQEEL SENDKKIKAL NHE IEELEK RKDKETGGIL RSLE DALAE AQRVNTKSQS AFDLK KKNL ACEESKRKEL EKNMVE DSK TLAAKEKEVK KITDGLH AL QEASNKDAEA LAAAQQHF N AVSAGLSSNE DGAEATLAG QMMACKNDIS KAQTEAKQAQ MKLKHAQQE LKNKQAEVKK M DSGYRKDQ EALEAVKRLK EK LEAEMKK LNYEENKEES LLE KRRQLS RDIGRLKETY EALL ARFPN LRFAYKDPEK NWNRN CVKG LVASLISVKD TSATTA LEL VAGERLYNVV VDTEVTG KK LLERGELKRR YTIIPLNK I SARCIAPETL RVAQNLVGP DNVHVALSLV EYKPELQKAM EFVFGTTFV CDNMDNAKKV A FDKRIMTR TVTLGGDVFD PH GTLSGGA RSQAASILTK FQE LKDVQD ELRIKENELR ALEE ELAGL KNTAEKYRQL KQQWE MKTE EADLLQTKLQ QSSYHK QQE ELDALKKTIE ESEETLK NT KEIQRKAEEK YEVLENKM K NAEAEREREL KDAQKKLDC AKTKADASSK KMKEKQQEVE AITLELEEL KREHTSYKQQ L EAVNEAIK SYESQIEVMA AE VAKNKES VNKAQEEVTK QKE VITAQD TVIKAKYAEV AKHK EQNND SQLKIKELDH NISKH KREA EDGAAKVSKM LKDYDW INA ERHLFGQPNS AYDFKTN NP KEAGQRLQKL QEMKEKLG R NVNMRAMNVL TEAEERYND LMKKKRIVEN DKSKILTTIE DLDQKKNQA LNIAWQKVNK D FGSIFSTL LPGANAMLAP PE GQTVLDG LEFKVALGNT WKE NLTELS GGQRSLVALS LILS MLLFK PAPIYILDEV DAALD LSHT QNIGQMLRTH FTHSQF IVV SLKEGMFNNA NVLFKTK FV DGVSTVARFT QCQNGKIS K EAKSKAKPPK GAHVEV |
-Macromolecule #2: SMC4
| Macromolecule | Name: SMC4 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAHHHHHHHH HHLEVLFQGP RKGTQPSTA RRREEGPPPP S PDGASSDA EPEPPSGRTE SP ATAAETA SEELDNRSLE EIL NSIPPP PPPAMTNEAG APRL MITHI VNQNFKSYAG EKILG PFHK RFSCIIGPNG SGKSNV IDS MLFVFGYRAQ KIRSKKL SV ...String: MAHHHHHHHH HHLEVLFQGP RKGTQPSTA RRREEGPPPP S PDGASSDA EPEPPSGRTE SP ATAAETA SEELDNRSLE EIL NSIPPP PPPAMTNEAG APRL MITHI VNQNFKSYAG EKILG PFHK RFSCIIGPNG SGKSNV IDS MLFVFGYRAQ KIRSKKL SV LIHNSDEHKD IQSCTVEV H FQKIIDKEGD DYEVIPNSN FYVSRTACRD NTSVYHISGK KKTFKDVGN LLRSHGIDLD H NRFLILQG EVEQIAMMKP KG QTEHDEG MLEYLEDIIG CGR LNEPIK VLCRRVEILN EHRG EKLNR VKMVEKEKDA LEGEK NIAI EFLTLENEIF RKKNHV CQY YIYELQKRIA EMETQKE KI HEDTKEINEK SNILSNEM K AKNKDVKDTE KKLNKITKF IEENKEKFTQ LDLEDVQVRE KLKHATSKA KKLEKQLQKD K EKVEEFKS IPAKSNNIIN ET TTRNNAL EKEKEKEEKK LKE VMDSLK QETQGLQKEK ESRE KELMG FSKSVNEARS KMDVA QSEL DIYLSRHNTA VSQLTK AKE ALIAASETLK ERKAAIR DI EGKLPQTEQE LKEKEKEL Q KLTQEETNFK SLVHDLFQK VEEAKSSLAM NRSRGKVLDA IIQEKKSGR IPGIYGRLGD L GAIDEKYD VAISSCCHAL DY IVVDSID IAQECVNFLK RQN IGVATF IGLDKMAVWA KKMT EIQTP ENTPRLFDLV KVKDE KIRQ AFYFALRDTL VADNLD QAT RVAYQKDRRW RVVTLQG QI IEQSGTMTGG GSKVMKGR M GSSLVIEISE EEVNKMESQ LQNDSKKAMQ IQEQKVQLEE RVVKLRHSE REMRNTLEKF T ASIQRLIE QEEYLNVQVK EL EANVLAT APDKKKQKLL EEN VSAFKT EYDAVAEKAG KVEA EVKRL HNTIVEINNH KLKAQ QDKL DKINKQLDEC ASAITK AQV AIKTADRNLQ KAQDSVL RT EKEIKDTEKE VDDLTAEL K SLEDKAAEVV KNTNAAEES LPEIQKEHRN LLQELKVIQE NEHALQKDA LSIKLKLEQI D GHIAEHNS KIKYWHKEIS KI SLHPIED NPIEEISVLS PED LEAIKN PDSITNQIAL LEAR CHEMK PNLGAIAEYK KKEEL YLQR VAELDKITYE RDSFRQ AYE DLRKQRLNEF MAGFYII TN KLKENYQMLT LGGDAELE L VDSLDPFSEG IMFSVRPPK KSWKKIFNLS GGEKTLSSLA LVFALHHYK PTPLYFMDEI D AALDFKNV SIVAFYIYEQ TK NAQFIII SLRNNMFEIS DRL IGIYKT YNITKSVAVN PKEI ASKGL C |
-Macromolecule #3: CapH2
| Macromolecule | Name: CapH2 / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: EDVEARFAHL LQPIRDLTK NWEVDVAAQL GEYLEELDQI CISFDEGKT TMNFIEAALL I QGSACVYS KKVEYLYSLV YQ ALDFISG KRRAKQLSSV QED RANGVA SSGVPQEAEN EFLS LDDFP DSRTNVDLKN DQTPS EVLI IPLLPMALVA PDEMEK NNN ...String: EDVEARFAHL LQPIRDLTK NWEVDVAAQL GEYLEELDQI CISFDEGKT TMNFIEAALL I QGSACVYS KKVEYLYSLV YQ ALDFISG KRRAKQLSSV QED RANGVA SSGVPQEAEN EFLS LDDFP DSRTNVDLKN DQTPS EVLI IPLLPMALVA PDEMEK NNN PLYSRQGEVL ASRKDFR MN TCVPHPRGAF MLEPEGMS P MEPAGVSPMP GTQKDTGRT EEQPMEVSVC RSPVPALGFS QEPGPSPEG PMPLGGGEDE D AEEAVELP EASAPKAALE PK ESRSPQQ SAALPRRYML RER EGAPEP ASCVKETPDP WQSL DPFDS LESKPFKKGR PYSVP PCVE EALGQKRKRK GAAKLQ DFH QWYLAAYADH ADSRRLR RK GPSFADMEVL YWTHVKEQ L ETLRKLQRRE VAEQWLRPA EEDHLEDSLE DLGAADDFLE PEEYMEPEG ADPREAADLD A VPMSLSYE ELVRRNVELF IA TSQKFVQ ETELSQRIRD WED TVQPLL QEQEQHVPFD IHTY GDQLV SRFPQLNEWC PFAEL VAGQ PAFEVCRSML ASLQLA NDY TVEITQQPGL EMAVDTM SL RLLTHQRAHK RFQTYAAP S MAQPENLYFQ SWSHPQFEK GGGSGGGSGG GSWSHPQFEK |
-Macromolecule #4: CapD3
| Macromolecule | Name: CapD3 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: VALRGLGSGL QPWCPLDLR LEWVDTVWEL DFTETEPLDP SIEAEIIET GLAAFTKLYE S LLPFATGE HGSMESIWTF FI ENNVSHS TLVALFYHFV QIV HKKNVS VQYREYGLHA AGLY FLLLE VPGSVANQVF HPVMF DKCI QTLKKSWPQE SNLNRK RKK ...String: VALRGLGSGL QPWCPLDLR LEWVDTVWEL DFTETEPLDP SIEAEIIET GLAAFTKLYE S LLPFATGE HGSMESIWTF FI ENNVSHS TLVALFYHFV QIV HKKNVS VQYREYGLHA AGLY FLLLE VPGSVANQVF HPVMF DKCI QTLKKSWPQE SNLNRK RKK EQPKSSQANP GRHRKRG KP PRREDIEMDE IIEEQEDE N ICFSARDLSQ IRNAIFHLL KNFLRLLPKF SLKEKPQCVQ NCIEVFVSL TNFEPVLHEC H VTQARALN QAKYIPELAY YG LYLLCSP IHGEGDKVIS CVF HQMLSV ILMLEVGEGS HRAP LAVTS QVINCRNQAV QFISA LVDE LKESIFPVVR ILLQHI CAK VVDKSEYRTF AAQSLVQ LL SKLPCGEYAM FIAWLYKY S RSSKIPHRVF TLDVVLALL ELPEREVDNT LSLEHQKFLK HKFLVQEIM FDRCLDKAPT V RSKALSSF AHCLELTVTS AS ESILELL INSPTFSVIE SHP GTLLRN SSAFSYQRQT SNRS EPSGE INIDSSGETV GSGER CVMA MLRRRIRDEK TNVRKS ALQ VLVSILKHCD VSGMKED LW ILQDQCRDPA VSVRKQAL Q SLTELLMAQP RCVQIQKAW LRGVVPVVMD CESTVQEKAL EFLDQLLLQ NIRHHSHFHS G DDSQVLAW ALLTLLTTES QE LSRYLNK AFHIWSKKEK FSP TFINNV ISHTGTEHSA PAWM LLSKI AGSSPRLDYS RIIQS WEKI SSQQNPNSNT LGHILC VIG HIAKHLPKST RDKVTDA VK CKLNGFQWSL EVISSAVD A LQRLCRASAE TPAEEQELL TQVCGDVLST CEHRLSNIVL KENGTGNMD EDLLVKYIFT L GDIAQLCP ARVEKRIFLL IQ SVLASSA DADHSPSSQG SSE APASQP PPQVRGSVMP SVIR AHAII TLGKLCLQHE DLAKK SIPA LVRELEVCED VAVRNN VII VMCDLCIRYT IMVDKYI PN ISMCLKDSDP FIRKQTLI L LTNLLQEEFV KWKGSLFFR FVSTLIDSHP DIASFGEFCL AHLLLKRNP VMFFQHFIEC I FHFNNYEK HEKYNKFPQS ER EKRLFSL KGKSNKERRM KIY KFLLEH FTDEQRFNIT SKIC LSILA CFADGILPLD LDASE LLSD TFEVLSSKEI KLLAMR SKP DKDLLMEEDD MALANVV MQ EAQKKLISQV QKRNFIEN I IPIIISLKTV LEKNKIPAL RELMHYLREV MQDYRDELKD FFAVDKQLA SELEYDMKKY Q EQLVQEQE LAKHADVAGT AG GAEVAPV AQVALCLETV PVP AGQENP AMSPAVSQPC TPRA SAGHV AVSSPTPETG PLQRL LPKA RPMSLSTIAI LNSVKK AVE SKSRHRSRSL GVLPFTL NS GSPEKTCSQV SSYSLEQE S NGEIEHVTKR AISTPEKSI SDVTFGAGVS YIGTPRTPSS AKEKIEGRS QGNDILCLSL P DKPPPQPQ QWNVRSPARN KD TPACSRR SLRKTPLKTA N |
-Macromolecule #5: CapG2
| Macromolecule | Name: CapG2 / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MEKRETFVQA VSKELVGEFL QFVQLDKEA SDPFSLNELL D ELSRKQKE ELWQRLKNLL TD VLLESPV DGWQVVEAQG EDN METEHG SKMRKSIEII YAIT SVILA SVSVINESEN YEALL ECVI ILNGILYALP ESERKL QSS IQDLCVTWWE KGLPAKE DT ...String: MEKRETFVQA VSKELVGEFL QFVQLDKEA SDPFSLNELL D ELSRKQKE ELWQRLKNLL TD VLLESPV DGWQVVEAQG EDN METEHG SKMRKSIEII YAIT SVILA SVSVINESEN YEALL ECVI ILNGILYALP ESERKL QSS IQDLCVTWWE KGLPAKE DT GKTAFVMLLR RSLETKTG A DVCRLWRIHQ ALYCFDYDL EESGEIKDML LECFININYI KKEEGRRFL SCLFNWNINF I KMIHGTIK NQLQGLQKSL MV YIAEIYF RAWKKASGKI LEA IENDCI QDFMFHGIHL PRRS PVHSK VREVLSYFHH QKKVR QGVE EMLYRLYKPI LWRGLK ARN SEVRSNAALL FVEAFPI RD PNLHAIEMDS EIQKQFEE L YSLLEDPYPM VRSTGILGV CKITSKYWEM MPPTILIDLL KKVTGELAF DTSSADVRCS V FKCLPMIL DNKLSHPLLE QL LPALRYS LHDNSEKVRV AFV DMLLKI KAVRAAKFWK ICPM EHILV RLETDSRPVS RRLVS LIFN SFLPVNQPEE VWCERC VTL VQMNHAAARR FYQYAHE HT ACTNIAKLIH VIRHCLNA C IQRAVREPPE DEEEEDGRE KENVTVLDKT LSVNDVACMA GLLEIIVIL WKSIDRSMEN N KEAKLYTI NKFASVLPEY LK VFKDDRC KIPLFMLMSF MPA SAVPPF SCGVISTLRS REEG AVDKS YCTLLDCLCS WGQVG HILE LVDNWLPTEH AQAKSN TAS KGRVQIHDTR PVKPELA LV YIEYLLTHPK NRECLLSA P RKKLNHLLKA LETSKADLE SLLQTPGGKP RGFSEAAAPR AFGLHCRLS IHLQHKFCSE G KVYLSMLE DTGFWLESKI LS FIQDQEE DYLKLHRVIY QQI IQTYLT VCKDVVMVGL GDHQ FQMQL LQRSLGIMQT VKGFF YVSL LLDILKEITG SSLIQK TDS DEEVAMLLDT VQKVFQK ML ECIARSFRKQ PEEGLRLL Y SVQRPLHEFI TAVQSRHTD TPVHRGVLST LIAGPVVEIS HQLRKVSDV EELTPPEHLS D LPPFSRCL IGIIIKSSNV VR SFLDELK ACVASNDIEG IVC LTAAVH IILVINAGKH KSSK VREVA ATVHRKLKTF MEITL EEDS IERFLYESSS RTLGEL LNS |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
Details: Grafix crosslinked sample was gel filtered on a superose 6 column into EM buffer (20 mM HEPES [pH 8], 200 mM KCl) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Acetate Details: The grids were glow discharged for 1 minute at 15 mA, the sample applied for 10 seconds and then washed twice with water before staining with 2% uranyl acetate for 1 minute before blotting and drying. | |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)