[English] 日本語
 Yorodumi
Yorodumi- EMDB-10755: Cryo-EM structure of norovirus GII.4c VLP with T=4 icosahedral sy... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10755 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
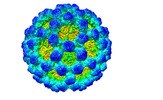
| Title | Cryo-EM structure of norovirus GII.4c VLP with T=4 icosahedral symmetry | |||||||||
 Map data Map data | Cryo-EM structure of GII.4c VLPs expressed in H5 insect cells with T=4 icosahedral symmetry | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Norovirus 06-AM-11/2006/GII.4/Yerseke/2006a Norovirus 06-AM-11/2006/GII.4/Yerseke/2006a | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.17 Å | |||||||||
 Authors Authors | Devant JM / Hansman GS | |||||||||
 Citation Citation |  Journal: Virology / Year: 2021 Journal: Virology / Year: 2021Title: Structural heterogeneity of a human norovirus vaccine candidate. Authors: Jessica M Devant / Grant S Hansman /  Abstract: Human norovirus virus-like particles (VLPs) are assumed to be morphologically and antigenically similar to virion particles. The norovirus virion is assembled from 180 copies of the capsid protein ...Human norovirus virus-like particles (VLPs) are assumed to be morphologically and antigenically similar to virion particles. The norovirus virion is assembled from 180 copies of the capsid protein (VP1) and exhibits T = 3 icosahedral symmetry. In this study, we showed that the vaccine candidate GII.4c VP1 formed T = 1 and T = 3 VLPs, but mainly assembled into T = 4 icosahedral particles that were composed of 240 VP1 copies. In contrast, another clinically important genotype, GII.17, almost exclusively folded into T = 3 VLPs. Interestingly, the GII.4c T = 1 particles had higher binding capacities to norovirus-specific Nanobodies than to GII.4c T = 3 and T = 4 particles. Our data indicated that the occluded Nanobody-binding epitopes on the T = 1 particles were more accessible compared to the larger T = 3 and T = 4 particles. Overall, this new data revealed that GII.4c VLPs had a preference for forming the T = 4 icosahedral symmetry and future studies with varied sized norovirus VLPs should take caution when examining antigenicity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10755.map.gz emd_10755.map.gz | 440.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10755-v30.xml emd-10755-v30.xml emd-10755.xml emd-10755.xml | 11.2 KB 11.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10755.png emd_10755.png | 252.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10755 http://ftp.pdbj.org/pub/emdb/structures/EMD-10755 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10755 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10755 | HTTPS FTP |
-Validation report
| Summary document |  emd_10755_validation.pdf.gz emd_10755_validation.pdf.gz | 228.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10755_full_validation.pdf.gz emd_10755_full_validation.pdf.gz | 227.3 KB | Display | |
| Data in XML |  emd_10755_validation.xml.gz emd_10755_validation.xml.gz | 7.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10755 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10755 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10755 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10755 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10755.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10755.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of GII.4c VLPs expressed in H5 insect cells with T=4 icosahedral symmetry | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.375 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Norovirus 06-AM-11/2006/GII.4/Yerseke/2006a
| Entire | Name:  Norovirus 06-AM-11/2006/GII.4/Yerseke/2006a Norovirus 06-AM-11/2006/GII.4/Yerseke/2006a |
|---|---|
| Components |
|
-Supramolecule #1: Norovirus 06-AM-11/2006/GII.4/Yerseke/2006a
| Supramolecule | Name: Norovirus 06-AM-11/2006/GII.4/Yerseke/2006a / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 1631306 Sci species name: Norovirus 06-AM-11/2006/GII.4/Yerseke/2006a Sci species strain: GII.4c / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Host system | Organism:  Trichoplusia ni (cabbage looper) / Recombinant strain: HighFive insect cells Trichoplusia ni (cabbage looper) / Recombinant strain: HighFive insect cells |
| Virus shell | Shell ID: 1 / Diameter: 520.0 Å / T number (triangulation number): 4 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Name: PBS / Details: PBS buffer |
| Grid | Model: Quantifoil R0.6/1 / Material: COPPER/RHODIUM / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 285 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 2 / Number real images: 3983 / Average electron dose: 15.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Software - Name: CTFFIND (ver. 4) |
|---|---|
| Final reconstruction | Number classes used: 1 / Applied symmetry - Point group: I (icosahedral) / Resolution.type: BY AUTHOR / Resolution: 4.17 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 2.4.0) / Number images used: 118971 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 2.4.0) |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cryoSPARC (ver. 2.4.0) |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















