+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1075 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Molecular architecture of the prolate head of bacteriophage T4. | |||||||||
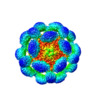
 Map data Map data | cryoEM reconstruction of the bacteriophage T4 head | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 22.5 Å | |||||||||
 Authors Authors | Fokine A / Chipman PR / Leiman PG / Mesyanzhinov VV / Rao VB / Rossmann MG | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2004 Journal: Proc Natl Acad Sci U S A / Year: 2004Title: Molecular architecture of the prolate head of bacteriophage T4. Authors: Andrei Fokine / Paul R Chipman / Petr G Leiman / Vadim V Mesyanzhinov / Venigalla B Rao / Michael G Rossmann /  Abstract: The head of bacteriophage T4 is a prolate icosahedron with one unique portal vertex to which the phage tail is attached. The three-dimensional structure of mature bacteriophage T4 head has been ...The head of bacteriophage T4 is a prolate icosahedron with one unique portal vertex to which the phage tail is attached. The three-dimensional structure of mature bacteriophage T4 head has been determined to 22-A resolution by using cryo-electron microscopy. The T4 capsid has a hexagonal surface lattice characterized by the triangulation numbers T(end) = 13 laevo for the icosahedral caps and T(mid) = 20 for the midsection. Hexamers of the major capsid protein gene product (gp)23* and pentamers of the vertex protein gp24*, as well as the outer surface proteins highly antigenic outer capsid protein (hoc) and small outer capsid protein (soc), are clearly evident in the reconstruction. The size and shape of the gp23* hexamers are similar to the major capsid protein organization of bacteriophage HK97. The binding sites and shape of the hoc and soc proteins have been established by analysis of the soc(-) and hoc(-)soc(-) T4 structures. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1075.map.gz emd_1075.map.gz | 22.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1075-v30.xml emd-1075-v30.xml emd-1075.xml emd-1075.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| Images |  1075.gif 1075.gif 400_1075.gif 400_1075.gif 400_10750.gif 400_10750.gif 400_10751.gif 400_10751.gif 400_10752.gif 400_10752.gif 400_10753.gif 400_10753.gif 400_10754.gif 400_10754.gif 400_10755.gif 400_10755.gif 400_10756.gif 400_10756.gif 400_10757.gif 400_10757.gif 400_10758.gif 400_10758.gif 400_10759.gif 400_10759.gif 80_1075.gif 80_1075.gif 80_10750.gif 80_10750.gif 80_10751.gif 80_10751.gif 80_10752.gif 80_10752.gif 80_10753.gif 80_10753.gif 80_10754.gif 80_10754.gif 80_10755.gif 80_10755.gif 80_10756.gif 80_10756.gif 80_10757.gif 80_10757.gif 80_10758.gif 80_10758.gif 80_10759.gif 80_10759.gif emd_1075.tif emd_1075.tif | 74.2 KB 61.4 KB 62.7 KB 55.9 KB 37.8 KB 29.4 KB 90.3 KB 106.2 KB 72.4 KB 51.9 KB 104.2 KB 76.2 KB 2.7 KB 4.3 KB 3.7 KB 2.9 KB 2.8 KB 5.4 KB 5.9 KB 4.5 KB 3.5 KB 5.9 KB 4.8 KB 737.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1075 http://ftp.pdbj.org/pub/emdb/structures/EMD-1075 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1075 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1075 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_1075.map.gz / Format: CCP4 / Size: 27.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_1075.map.gz / Format: CCP4 / Size: 27.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoEM reconstruction of the bacteriophage T4 head | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.96 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : bacteriophage T4 prolate head
| Entire | Name: bacteriophage T4 prolate head |
|---|---|
| Components |
|
-Supramolecule #1000: bacteriophage T4 prolate head
| Supramolecule | Name: bacteriophage T4 prolate head / type: sample / ID: 1000 Details: Sample was the native bacteriophage T4. Only the head part of the phage was reconstructed. Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 194 MDa / Method: STEM, 194 MDa is the mass of the DNA filled head. |
-Supramolecule #1: Enterobacteria phage T4
| Supramolecule | Name: Enterobacteria phage T4 / type: virus / ID: 1 / Name.synonym: phage T4 / NCBI-ID: 10665 / Sci species name: Enterobacteria phage T4 / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No / Syn species name: phage T4 |
|---|---|
| Host (natural) | Organism: Ecoli / synonym: BACTERIA(EUBACTERIA) |
| Molecular weight | Theoretical: 194 MDa |
| Virus shell | Shell ID: 1 / Name: capsid width is 860 A, length is 1195 A |
| Virus shell | Shell ID: 2 / Name: Tmid=20; Tend=13 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.2 Details: Distilled water. The concentration of the virus particles was 10E11 particles pre ml. |
|---|---|
| Grid | Details: quantifoil copper grid 2um holes |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 90 K / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: in-house, gravity driven plunger Method: 3.5ul of sample hand blotted approx. 1sec |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM300FEG/T |
|---|---|
| Temperature | Average: 97 K |
| Alignment procedure | Legacy - Astigmatism: live fft at 190,000x |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 55 / Average electron dose: 29 e/Å2 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 47000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.1 µm / Nominal magnification: 45000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C5 (5 fold cyclic) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 22.5 Å / Resolution method: FSC 0.33 CUT-OFF / Software - Name: SPIDER Details: The reconstruction was performed by imposing 5-fold symmetry averaging. The 5-fold axis is along Z. Number images used: 5140 |
| Final angle assignment | Details: The reconstruction was made in SPIDER using the projection matching protocol. The reference projections were calculated in the interval theta: 0-90 degrees, phi: 0-72 degrees. |
-Atomic model buiding 1
| Details | Nothing to fit |
|---|
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















