+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10492 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Vip3Aa protoxin structure | ||||||||||||||||||
 Map data Map data | Half map 2 | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Vip3Aa / toxin / protoxin / beta prism / insecticidal protein / Vip3 | ||||||||||||||||||
| Function / homology | Vegetative insecticide protein 3 / Vegetative insecticide protein 3A N terminal / Carbohydrate-binding, CenC-like / Carbohydrate binding domain / hydrolase activity, acting on glycosyl bonds / Galactose-binding-like domain superfamily / Vegetative insecticidal protein Function and homology information Function and homology information | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
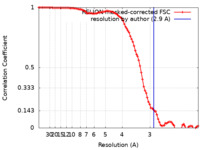
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | ||||||||||||||||||
 Authors Authors | Nunez-Ramirez R / Huesa J | ||||||||||||||||||
| Funding support |  Spain, 5 items Spain, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Molecular architecture and activation of the insecticidal protein Vip3Aa from Bacillus thuringiensis. Authors: Rafael Núñez-Ramírez / Juanjo Huesa / Yolanda Bel / Juan Ferré / Patricia Casino / Ernesto Arias-Palomo /  Abstract: Bacillus thuringiensis Vip3 (Vegetative Insecticidal Protein 3) toxins are widely used in biotech crops to control Lepidopteran pests. These proteins are produced as inactive protoxins that need to ...Bacillus thuringiensis Vip3 (Vegetative Insecticidal Protein 3) toxins are widely used in biotech crops to control Lepidopteran pests. These proteins are produced as inactive protoxins that need to be activated by midgut proteases to trigger cell death. However, little is known about their three-dimensional organization and activation mechanism at the molecular level. Here, we have determined the structures of the protoxin and the protease-activated state of Vip3Aa at 2.9 Å using cryo-electron microscopy. The reconstructions show that the protoxin assembles into a pyramid-shaped tetramer with the C-terminal domains exposed to the solvent and the N-terminal region folded into a spring-loaded apex that, after protease activation, drastically remodels into an extended needle by a mechanism akin to that of influenza haemagglutinin. These results provide the molecular basis for Vip3 activation and function, and serves as a strong foundation for the development of more efficient insecticidal proteins. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10492.map.gz emd_10492.map.gz | 7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10492-v30.xml emd-10492-v30.xml emd-10492.xml emd-10492.xml | 21.1 KB 21.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10492_fsc.xml emd_10492_fsc.xml | 9.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_10492.png emd_10492.png | 158.6 KB | ||
| Masks |  emd_10492_msk_1.map emd_10492_msk_1.map | 70.2 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10492.cif.gz emd-10492.cif.gz | 6.2 KB | ||
| Others |  emd_10492_additional_1.map.gz emd_10492_additional_1.map.gz emd_10492_additional_2.map.gz emd_10492_additional_2.map.gz emd_10492_half_map_1.map.gz emd_10492_half_map_1.map.gz emd_10492_half_map_2.map.gz emd_10492_half_map_2.map.gz | 53 MB 2.7 MB 53.3 MB 53.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10492 http://ftp.pdbj.org/pub/emdb/structures/EMD-10492 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10492 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10492 | HTTPS FTP |
-Related structure data
| Related structure data |  6tfjMC  6tfkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10492.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10492.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.048 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
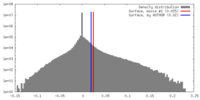
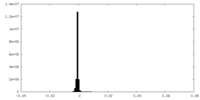
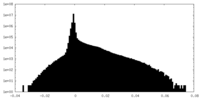
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10492_msk_1.map emd_10492_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened EM map of the Vip3Aa protoxin
| File | emd_10492_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened EM map of the Vip3Aa protoxin | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Focused classification map of the C-terminal domains of the protoxin
| File | emd_10492_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused classification map of the C-terminal domains of the protoxin | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_10492_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Focused classification map of the C-terminal domains of the protoxin
| File | emd_10492_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused classification map of the C-terminal domains of the protoxin | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Vip3Aa protoxin
| Entire | Name: Vip3Aa protoxin |
|---|---|
| Components |
|
-Supramolecule #1: Vip3Aa protoxin
| Supramolecule | Name: Vip3Aa protoxin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 350 KDa |
-Macromolecule #1: Vegetative insecticidal protein
| Macromolecule | Name: Vegetative insecticidal protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 88.762805 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNKNNTKLST RALPSFIDYF NGIYGFATGI KDIMNMIFKT DTGGDLTLDE ILKNQQLLND ISGKLDGVNG SLNDLIAQGN LNTELSKEI LKIANEQNQV LNDVNNKLDA INTMLRVYLP KLTSMLSDVM KQNYALSLQI EYLSKQLQEI SDKLDIINVN V LINSTLTE ...String: MNKNNTKLST RALPSFIDYF NGIYGFATGI KDIMNMIFKT DTGGDLTLDE ILKNQQLLND ISGKLDGVNG SLNDLIAQGN LNTELSKEI LKIANEQNQV LNDVNNKLDA INTMLRVYLP KLTSMLSDVM KQNYALSLQI EYLSKQLQEI SDKLDIINVN V LINSTLTE ITPAYQRIKY VNEKFEELTF ATETSSKVKK DGSPADILDE LTELTELAKS VTKNDVDGFE FYLNTFHDVM VG NNLFGRS ALKTASELIT KENVKTSGSE VGNVYNFLIV LTALQAKAFL TLTTCRKLLG LADIDYTSIM NEHLNKEKEE FRV NILPTL SNTFSNPNYA KVKGSDEDAK MIVEAKPGHA LIGFEISNDS ITVLKVYEAK LKQNYQVDKD SLSEVIYGDM DKLL CPDQS EQIYYTNNIV FPNEYVITKI DFTKKMKTLR YEVTANFYDS STGEIDLNKK KVESSEAEYR TLSANDDGVY MPLGV ISET FLTPINGFGL QADENSRLIT LTCKSYLREL LLATDLSNKE TKLIVPPSGF ISNIVENGSI EEDNLEPWKA NNKNAY VDH TGGVNGTKAL YVHKDGGISQ FIGDKLKPKT EYVIQYTVKG KPSIHLKDEN TGYIHYEDTN NNLEDYQTIN KRFTTGT DL KGVYLILKSQ NGDEAWGDNF IILEISPSEK LLSPELINTN NWTSTGSTNI SGNTLTLYQG GRGILKQNLQ LDSFSTYR V YFSVSGDANV RIRNSREVLF EKRYMSGAKD VSEMFTTKFE KDNFYIELSQ GNNLYGGPIV HFYDVSIK UniProtKB: Vegetative insecticidal protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 56.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | The atomic coordinates were manually modeled de novo in the cryo-EM map using Coot, and then subjected to iterative rounds of real space refinement using Phenix |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER |
| Output model |  PDB-6tfj: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)