+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10466 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-ET on GroEL | |||||||||
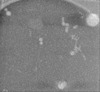
 Map data Map data | Reconstructed tomography data.The GroEL specimen preparation method is Preassis, and a Quantifoil continuous carbon film grid (R2/2) was used. In the hole of this tomogram, the carbon film was broken. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | electron tomography / cryo EM | |||||||||
 Authors Authors | Zou XD / Zhao JJ / Xu HY / Carroni M | |||||||||
| Funding support |  Sweden, 2 items Sweden, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: A simple pressure-assisted method for MicroED specimen preparation. Authors: Jingjing Zhao / Hongyi Xu / Hugo Lebrette / Marta Carroni / Helena Taberman / Martin Högbom / Xiaodong Zou /   Abstract: Micro-crystal electron diffraction (MicroED) has shown great potential for structure determination of macromolecular crystals too small for X-ray diffraction. However, specimen preparation remains a ...Micro-crystal electron diffraction (MicroED) has shown great potential for structure determination of macromolecular crystals too small for X-ray diffraction. However, specimen preparation remains a major bottleneck. Here, we report a simple method for preparing MicroED specimens, named Preassis, in which excess liquid is removed through an EM grid with the assistance of pressure. We show the ice thicknesses can be controlled by tuning the pressure in combination with EM grids with appropriate carbon hole sizes. Importantly, Preassis can handle a wide range of protein crystals grown in various buffer conditions including those with high viscosity, as well as samples with low crystal concentrations. Preassis is a simple and universal method for MicroED specimen preparation, and will significantly broaden the applications of MicroED. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10466.map.gz emd_10466.map.gz | 210.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10466-v30.xml emd-10466-v30.xml emd-10466.xml emd-10466.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10466.png emd_10466.png | 126.8 KB | ||
| Others |  emd_10466_additional_1.map.gz emd_10466_additional_1.map.gz emd_10466_additional_2.map.gz emd_10466_additional_2.map.gz emd_10466_additional_3.map.gz emd_10466_additional_3.map.gz | 80.9 MB 778.5 MB 774.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10466 http://ftp.pdbj.org/pub/emdb/structures/EMD-10466 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10466 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10466 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10466.map.gz / Format: CCP4 / Size: 297.4 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) Download / File: emd_10466.map.gz / Format: CCP4 / Size: 297.4 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstructed tomography data.The GroEL specimen preparation method is Preassis, and a Quantifoil continuous carbon film grid (R2/2) was used. In the hole of this tomogram, the carbon film was broken. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 13.8 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Reconstructed tomography data.The GroEL specimen preparation method is...
| File | emd_10466_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstructed tomography data.The GroEL specimen preparation method is Preassis, and a Quantifoil continuous carbon film grid (R2/2) was used. In the hole of this tomogram, the carbon film was intact. | ||||||||||||
| Projections & Slices |
| ||||||||||||
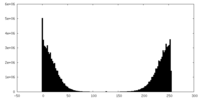
| Density Histograms |
-Additional map: Raw tomography data. The GroEL specimen preparation method...
| File | emd_10466_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw tomography data. The GroEL specimen preparation method is Preassis, and a Quantifoil continuous carbon film grid (R2/2) was used. In the hole of this tomogram, the carbon film was intact. | ||||||||||||
| Projections & Slices |
| ||||||||||||
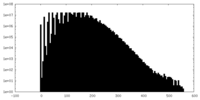
| Density Histograms |
-Additional map: Raw tomography data. The GroEL specimen preparation method...
| File | emd_10466_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Raw tomography data. The GroEL specimen preparation method is Preassis, and a Quantifoil continuous carbon film grid (R2/2) was used. In the hole of this tomogram, the carbon film was broken. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : GroEL
| Entire | Name: GroEL |
|---|---|
| Components |
|
-Supramolecule #1: GroEL
| Supramolecule | Name: GroEL / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 0.58 kDa/nm |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.70 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 0.2 nm / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Details: A new freezing method was used, called Preassis. |
| Details | The GroEL grid for cryoET data collection was prepared by a new specimen preparation method, called Preassis. A Quantifoil continuous carbon grid was used. |
| Sectioning | Other: NO SECTIONING |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number images used: 50 |
|---|
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)