[English] 日本語
 Yorodumi
Yorodumi- EMDB-10258: Cryo-EM structure of nanodisc reconstituted yeast ALG6 in complex... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10258 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of nanodisc reconstituted yeast ALG6 in complex with 6AG9 Fab | ||||||||||||
 Map data Map data | Nanodisc reconstituted yeast ALG6 in complex with 6AG9 Fab | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Glycosyltransferase / Glucosyltransferase / GT-C / N-Glycosylation / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationdolichyl-P-Glc:Man9GlcNAc2-PP-dolichol alpha-1,3-glucosyltransferase / dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase activity / Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein / dolichol-linked oligosaccharide biosynthetic process / hexosyltransferase activity / : / aerobic respiration / endoplasmic reticulum membrane / endoplasmic reticulum Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||
 Authors Authors | Bloch JS / Pesciullesi G | ||||||||||||
| Funding support |  Switzerland, 3 items Switzerland, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Structure and mechanism of the ER-based glucosyltransferase ALG6. Authors: Joël S Bloch / Giorgio Pesciullesi / Jérémy Boilevin / Kamil Nosol / Rossitza N Irobalieva / Tamis Darbre / Markus Aebi / Anthony A Kossiakoff / Jean-Louis Reymond / Kaspar P Locher /   Abstract: In eukaryotic protein N-glycosylation, a series of glycosyltransferases catalyse the biosynthesis of a dolichylpyrophosphate-linked oligosaccharide before its transfer onto acceptor proteins. The ...In eukaryotic protein N-glycosylation, a series of glycosyltransferases catalyse the biosynthesis of a dolichylpyrophosphate-linked oligosaccharide before its transfer onto acceptor proteins. The final seven steps occur in the lumen of the endoplasmic reticulum (ER) and require dolichylphosphate-activated mannose and glucose as donor substrates. The responsible enzymes-ALG3, ALG9, ALG12, ALG6, ALG8 and ALG10-are glycosyltransferases of the C-superfamily (GT-Cs), which are loosely defined as containing membrane-spanning helices and processing an isoprenoid-linked carbohydrate donor substrate. Here we present the cryo-electron microscopy structure of yeast ALG6 at 3.0 Å resolution, which reveals a previously undescribed transmembrane protein fold. Comparison with reported GT-C structures suggests that GT-C enzymes contain a modular architecture with a conserved module and a variable module, each with distinct functional roles. We used synthetic analogues of dolichylphosphate-linked and dolichylpyrophosphate-linked sugars and enzymatic glycan extension to generate donor and acceptor substrates using purified enzymes of the ALG pathway to recapitulate the activity of ALG6 in vitro. A second cryo-electron microscopy structure of ALG6 bound to an analogue of dolichylphosphate-glucose at 3.9 Å resolution revealed the active site of the enzyme. Functional analysis of ALG6 variants identified a catalytic aspartate residue that probably acts as a general base. This residue is conserved in the GT-C superfamily. Our results define the architecture of ER-luminal GT-C enzymes and provide a structural basis for understanding their catalytic mechanisms. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10258.map.gz emd_10258.map.gz | 96.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10258-v30.xml emd-10258-v30.xml emd-10258.xml emd-10258.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
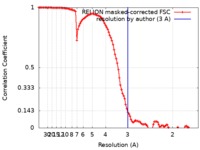
| FSC (resolution estimation) |  emd_10258_fsc.xml emd_10258_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_10258.png emd_10258.png | 110.3 KB | ||
| Filedesc metadata |  emd-10258.cif.gz emd-10258.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10258 http://ftp.pdbj.org/pub/emdb/structures/EMD-10258 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10258 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10258 | HTTPS FTP |
-Validation report
| Summary document |  emd_10258_validation.pdf.gz emd_10258_validation.pdf.gz | 548.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10258_full_validation.pdf.gz emd_10258_full_validation.pdf.gz | 548.4 KB | Display | |
| Data in XML |  emd_10258_validation.xml.gz emd_10258_validation.xml.gz | 11.6 KB | Display | |
| Data in CIF |  emd_10258_validation.cif.gz emd_10258_validation.cif.gz | 15.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10258 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10258 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10258 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10258 | HTTPS FTP |
-Related structure data
| Related structure data |  6sniMC  6snhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10429 (Title: 3 Å resolution single particle reconstruction of glucosyltransferase ALG6 in nanodisc EMPIAR-10429 (Title: 3 Å resolution single particle reconstruction of glucosyltransferase ALG6 in nanodiscData size: 2.8 TB Data #1: Motion corrected micrographs of the Alg6-Fab complex [micrographs - single frame] Data #2: Unaligned multi-frame micrographs of the Alg6-Fab complex [micrographs - multiframe] Data #3: Unaligned multi-frame micrographs of the Alg6-Fab complex [micrographs - multiframe] Data #4: Unaligned multi-frame micrographs of the Alg6-Fab complex [micrographs - multiframe] Data #5: Unaligned multi-frame micrographs of the Alg6-Fab complex [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10258.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10258.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Nanodisc reconstituted yeast ALG6 in complex with 6AG9 Fab | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Nanodisc reconstituted yeast ALG6 in complex with 6AG9 Fab
| Entire | Name: Nanodisc reconstituted yeast ALG6 in complex with 6AG9 Fab |
|---|---|
| Components |
|
-Supramolecule #1: Nanodisc reconstituted yeast ALG6 in complex with 6AG9 Fab
| Supramolecule | Name: Nanodisc reconstituted yeast ALG6 in complex with 6AG9 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Theoretical: 112.89472 KDa |
-Supramolecule #2: ALG6
| Supramolecule | Name: ALG6 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: 6AG9 Fab
| Supramolecule | Name: 6AG9 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: Dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase
| Macromolecule | Name: Dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: dolichyl-P-Glc:Man9GlcNAc2-PP-dolichol alpha-1,3-glucosyltransferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 64.423223 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GPGGSTGRLA GAGGEFVDMA IGKRLLVNKP AEESFYASPM YDFLYPFRPV GNQWLPEYII FVCAVILRCT IGLGPYSGKG SPPLYGDFE AQRHWMEITQ HLPLSKWYWY DLQYWGLDYP PLTAFHSYLL GLIGSFFNPS WFALEKSRGF ESPDNGLKTY M RSTVIISD ...String: GPGGSTGRLA GAGGEFVDMA IGKRLLVNKP AEESFYASPM YDFLYPFRPV GNQWLPEYII FVCAVILRCT IGLGPYSGKG SPPLYGDFE AQRHWMEITQ HLPLSKWYWY DLQYWGLDYP PLTAFHSYLL GLIGSFFNPS WFALEKSRGF ESPDNGLKTY M RSTVIISD ILFYFPAVIY FTKWLGRYRN QSPIGQSIAA SAILFQPSLM LIDHGHFQYN SVMLGLTAYA INNLLDEYYA MA AVCFVLS ICFKQMALYY APIFFAYLLS RSLLFPKFNI ARLTVIAFAT LATFAIIFAP LYFLGGGLKN IHQCIHRIFP FAR GIFEDK VANFWCVTNV FVKYKERFTI QQLQLYSLIA TVIGFLPAMI MTLLHPKKHL LPYVLIACSM SFFLFSFQVH EKTI LIPLL PITLLYSSTD WNVLSLVSWI NNVALFTLWP LLKKDGLHLQ YAVSFLLSNW LIGNFSFITP RFLPKSLTPG PSISS INSD YRRRSLLPYN VVWKSFIIGT YIAMGFYHFL DQFVAPPSKY PDLWVLLNCA VGFICFSIFW LWSYYKIFTS GSKSMK DL UniProtKB: Dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase |
-Macromolecule #2: 6AG9-Fab heavy chain
| Macromolecule | Name: 6AG9-Fab heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 24.953664 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVYSSSIHWV RQAPGKGLEW VASISSYSGY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC AREYWSWYSY SYGIDYWGQG TLVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF P EPVTVSWN ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NVYSSSIHWV RQAPGKGLEW VASISSYSGY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC AREYWSWYSY SYGIDYWGQG TLVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF P EPVTVSWN SGALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDKKVEPKS CDKTHT |
-Macromolecule #3: 6AG9-Fab light chain
| Macromolecule | Name: 6AG9-Fab light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 23.65025 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSYWVGYPI TFGQGTKVEI KRTVAAPSVF IFPPSDSQLK SGTASVVCLL NNFYPREAKV QWKVDNALQS G NSQESVTE ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSYWVGYPI TFGQGTKVEI KRTVAAPSVF IFPPSDSQLK SGTASVVCLL NNFYPREAKV QWKVDNALQS G NSQESVTE QDSKDSTYSL SSTLTLSKAD YEKHKVYACE VTHQGLSSPV TKSFNRGEC |
-Macromolecule #4: PHOSPHATIDYLETHANOLAMINE
| Macromolecule | Name: PHOSPHATIDYLETHANOLAMINE / type: ligand / ID: 4 / Number of copies: 5 / Formula: PTY |
|---|---|
| Molecular weight | Theoretical: 734.039 Da |
| Chemical component information |  ChemComp-PTY: |
-Macromolecule #5: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 5 / Number of copies: 3 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 7 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 2.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)