+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10255 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | BamABCDE in MSP1D1 nanodisc ensemble 0-8 | |||||||||||||||
 Map data Map data | BamABCDE in MSP1D1 nanodisc ensemble 0-8 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Outer membrane / OMP / beta-barrel / folding / insertion / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationBam protein complex / Gram-negative-bacterium-type cell outer membrane assembly / protein insertion into membrane / cell outer membrane / protein-macromolecule adaptor activity / cell adhesion / response to antibiotic / cell surface / identical protein binding / membrane Similarity search - Function | |||||||||||||||
| Biological species |   | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.8 Å | |||||||||||||||
 Authors Authors | Iadanza MG / Ranson NA | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2020 Journal: Commun Biol / Year: 2020Title: Distortion of the bilayer and dynamics of the BAM complex in lipid nanodiscs. Authors: Matthew G Iadanza / Bob Schiffrin / Paul White / Matthew A Watson / Jim E Horne / Anna J Higgins / Antonio N Calabrese / David J Brockwell / Roman Tuma / Antreas C Kalli / Sheena E Radford / Neil A Ranson /   Abstract: The β-barrel assembly machinery (BAM) catalyses the folding and insertion of β-barrel outer membrane proteins (OMPs) into the outer membranes of Gram-negative bacteria by mechanisms that remain ...The β-barrel assembly machinery (BAM) catalyses the folding and insertion of β-barrel outer membrane proteins (OMPs) into the outer membranes of Gram-negative bacteria by mechanisms that remain unclear. Here, we present an ensemble of cryoEM structures of the E. coli BamABCDE (BAM) complex in lipid nanodiscs, determined using multi-body refinement techniques. These structures, supported by single-molecule FRET measurements, describe a range of motions in the BAM complex, mostly localised within the periplasmic region of the major subunit BamA. The β-barrel domain of BamA is in a 'lateral open' conformation in all of the determined structures, suggesting that this is the most energetically favourable species in this bilayer. Strikingly, the BAM-containing lipid nanodisc is deformed, especially around BAM's lateral gate. This distortion is also captured in molecular dynamics simulations, and provides direct structural evidence for the lipid 'disruptase' activity of BAM, suggested to be an important part of its functional mechanism. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10255.map.gz emd_10255.map.gz | 8.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10255-v30.xml emd-10255-v30.xml emd-10255.xml emd-10255.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10255_fsc.xml emd_10255_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_10255.png emd_10255.png | 76.5 KB | ||
| Filedesc metadata |  emd-10255.cif.gz emd-10255.cif.gz | 6.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10255 http://ftp.pdbj.org/pub/emdb/structures/EMD-10255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10255 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10255 | HTTPS FTP |
-Related structure data
| Related structure data |  6sn9MC  6smxC  6sn0C  6sn2C  6sn3C  6sn4C  6sn5C  6sn7C  6sn8C  6so7C  6so8C  6soaC  6sobC  6socC  6sogC  6sohC  6sojC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10255.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10255.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BamABCDE in MSP1D1 nanodisc ensemble 0-8 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
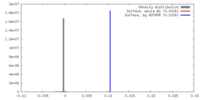
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Bam complex
| Entire | Name: Bam complex |
|---|---|
| Components |
|
-Supramolecule #1: Bam complex
| Supramolecule | Name: Bam complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: In MSP1D1 nanodisc with E. coli polar lipid extract |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: Outer membrane protein assembly factor BamA
| Macromolecule | Name: Outer membrane protein assembly factor BamA / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 87.783945 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: FVVKDIHFEG LQRVAVGAAL LSMPVRTGDT VNDEDISNTI RALFATGNFE DVRVLRDGDT LLVQVKERPT IASITFSGNK SVKDDMLKQ NLEASGVRVG ESLDRTTIAD IEKGLEDFYY SVGKYSASVK AVVTPLPRNR VDLKLVFQEG VSAEIQQINI V GNHAFTTD ...String: FVVKDIHFEG LQRVAVGAAL LSMPVRTGDT VNDEDISNTI RALFATGNFE DVRVLRDGDT LLVQVKERPT IASITFSGNK SVKDDMLKQ NLEASGVRVG ESLDRTTIAD IEKGLEDFYY SVGKYSASVK AVVTPLPRNR VDLKLVFQEG VSAEIQQINI V GNHAFTTD ELISHFQLRD EVPWWNVVGD RKYQKQKLAG DLETLRSYYL DRGYARFNID STQVSLTPDK KGIYVTVNIT EG DQYKLSG VEVSGNLAGH SAEIEQLTKI EPGELYNGTK VTKMEDDIKK LLGRYGYAYP RVQSMPEIND ADKTVKLRVN VDA GNRFYV RKIRFEGNDT SKDAVLRREM RQMEGAWLGS DLVDQGKERL NRLGFFETVD TDTQRVPGSP DQVDVVYKVK ERNT GSFNF GIGYGTESGV SFQAGVQQDN WLGTGYAVGI NGTKNDYQTY AELSVTNPYF TVDGVSLGGR LFYNDFQADD ADLSD YTNK SYGTDVTLGF PINEYNSLRA GLGYVHNSLS NMQPQVAMWR YLYSMGEHPS TSDQDNSFKT DDFTFNYGWT YNKLDR GYF PTDGSRVNLT GKVTIPGSDN EYYKVTLDTA TYVPIDDDHK WVVLGRTRWG YGDGLGGKEM PFYENFYAGG SSTVRGF QS NTIGPKAVYF PHQASNYDPD YDYECATQDG AKDLCKSDDA VGGNAMAVAS LEFITPTPFI SDKYANSVRT SFFWDMGT V WDTNWDSSQY SGYPDYSDPS NIRMSAGIAL QWMSPLGPLV FSYAQPFKKY DGDKAEQFQF NI UniProtKB: Outer membrane protein assembly factor BamA |
-Macromolecule #2: Outer membrane protein assembly factor BamB
| Macromolecule | Name: Outer membrane protein assembly factor BamB / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.692156 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: LFNSEEDVVK MSPLPTVENQ FTPTTAWSTS VGSGIGNFYS NLHPALADNV VYAADRAGLV KALNADDGKE IWSVSLAEKD GWFSKEPAL LSGGVTVSGG HVYIGSEKAQ VYALNTSDGT VAWQTKVAGE ALSRPVVSDG LVLIHTSNGQ LQALNEADGA V KWTVNLDM ...String: LFNSEEDVVK MSPLPTVENQ FTPTTAWSTS VGSGIGNFYS NLHPALADNV VYAADRAGLV KALNADDGKE IWSVSLAEKD GWFSKEPAL LSGGVTVSGG HVYIGSEKAQ VYALNTSDGT VAWQTKVAGE ALSRPVVSDG LVLIHTSNGQ LQALNEADGA V KWTVNLDM PSLSLRGESA PTTAFGAAVV GGDNGRVSAV LMEQGQMIWQ QRISQATGST EIDRLSDVDT TPVVVNGVVF AL AYNGNLT ALDLRSGQIM WKRELGSVND FIVDGNRIYL VDQNDRVMAL TIDGGVTLWT QSDLLHRLLT SPVLYNGNLV VGD SEGYLH WINVEDGRFV AQQKVDSSGF QTEPVAADGK LLIQAKDGTV YSITR UniProtKB: UNIPROTKB: A0A2S5ZNM3 |
-Macromolecule #3: Outer membrane protein assembly factor BamC
| Macromolecule | Name: Outer membrane protein assembly factor BamC / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 6.096815 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: CSSDSRYKRQ VSGDEAYLEA APLAELHAPA GMILPVTSGD YAIPVTNGSG AVGKALDIR UniProtKB: UNIPROTKB: A0A4T5IPK3 |
-Macromolecule #4: Outer membrane protein assembly factor BamD
| Macromolecule | Name: Outer membrane protein assembly factor BamD / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.008967 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVPDNPPNEI YATAQQKLQD GNWRQAITQL EALDNRYPFG PYSQQVQLDL IYAYYKNADL PLAQAAIDRF IRLNPTHPNI DYVMYMRGL TNMALDDSAL QGFFGVDRSD RDPQHARAAF SDFSKLVRGY PNSQYTTDAT KRLVFLKDRL AKYEYSVAEY Y TERGAWVA ...String: EVPDNPPNEI YATAQQKLQD GNWRQAITQL EALDNRYPFG PYSQQVQLDL IYAYYKNADL PLAQAAIDRF IRLNPTHPNI DYVMYMRGL TNMALDDSAL QGFFGVDRSD RDPQHARAAF SDFSKLVRGY PNSQYTTDAT KRLVFLKDRL AKYEYSVAEY Y TERGAWVA VVNRVEGMLR DYPDTQATRD ALPLMENAYR QMQMNAQAEK VAKIIAANSS UniProtKB: Outer membrane protein assembly factor BamD |
-Macromolecule #5: Outer membrane protein assembly factor BamE
| Macromolecule | Name: Outer membrane protein assembly factor BamE / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 9.728837 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ERVVYRPDIN QGNYLTANDV SKIRVGMTQQ QVAYALGTPL MSDPFGTNTW FYVFRQQPGH EGVTQQTLTL TFNSSGVLTN IDNKPAL UniProtKB: Outer membrane protein assembly factor BamE |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 281 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 4 / Number real images: 15504 / Average exposure time: 10.0 sec. / Average electron dose: 40.75 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.6 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)