[English] 日本語
 Yorodumi
Yorodumi- EMDB-10235: Cryo-electron microscopy structure of a RbcL-Raf1 supercomplex fr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10235 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron microscopy structure of a RbcL-Raf1 supercomplex from Synechococcus elongatus PCC 7942 | |||||||||||||||
 Map data Map data | Cryo-electron microscopy structure of a RbcL8-Raf18 supercomplex from the cyanobacterium Synechococcus elongatus 7942 | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationribulose bisphosphate carboxylase complex assembly / photorespiration / carboxysome / ribulose-bisphosphate carboxylase / ribulose-bisphosphate carboxylase activity / reductive pentose-phosphate cycle / carbon fixation / photosynthesis / monooxygenase activity / magnesium ion binding / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  Synechococcus elongatus PCC 7942 (bacteria) / Synechococcus elongatus PCC 7942 (bacteria) /  Synechococcus elongatus PCC 7942 = FACHB-805 (bacteria) / Synechococcus elongatus PCC 7942 = FACHB-805 (bacteria) /  Synechococcus elongatus (strain PCC 7942 / FACHB-805) (bacteria) Synechococcus elongatus (strain PCC 7942 / FACHB-805) (bacteria) | |||||||||||||||
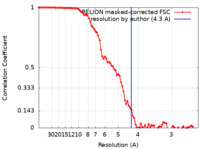
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||||||||
 Authors Authors | Huang F / Kong W-W / Sun Y / Chen T / Dykes GF / Jiang YL / Liu LN | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Rubisco accumulation factor 1 (Raf1) plays essential roles in mediating Rubisco assembly and carboxysome biogenesis. Authors: Fang Huang / Wen-Wen Kong / Yaqi Sun / Taiyu Chen / Gregory F Dykes / Yong-Liang Jiang / Lu-Ning Liu /   Abstract: Carboxysomes are membrane-free organelles for carbon assimilation in cyanobacteria. The carboxysome consists of a proteinaceous shell that structurally resembles virus capsids and internal enzymes ...Carboxysomes are membrane-free organelles for carbon assimilation in cyanobacteria. The carboxysome consists of a proteinaceous shell that structurally resembles virus capsids and internal enzymes including ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco), the primary carbon-fixing enzyme in photosynthesis. The formation of carboxysomes requires hierarchical self-assembly of thousands of protein subunits, initiated from Rubisco assembly and packaging to shell encapsulation. Here we study the role of Rubisco assembly factor 1 (Raf1) in Rubisco assembly and carboxysome formation in a model cyanobacterium, PCC7942 (Syn7942). Cryo-electron microscopy reveals that Raf1 facilitates Rubisco assembly by mediating RbcL dimer formation and dimer-dimer interactions. Syn7942 cells lacking Raf1 are unable to form canonical intact carboxysomes but generate a large number of intermediate assemblies comprising Rubisco, CcaA, CcmM, and CcmN without shell encapsulation and a low abundance of carboxysome-like structures with reduced dimensions and irregular shell shapes and internal organization. As a consequence, the Raf1-depleted cells exhibit reduced Rubisco content, CO-fixing activity, and cell growth. Our results provide mechanistic insight into the chaperone-assisted Rubisco assembly and biogenesis of carboxysomes. Advanced understanding of the biogenesis and stepwise formation process of the biogeochemically important organelle may inform strategies for heterologous engineering of functional CO-fixing modules to improve photosynthesis. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10235.map.gz emd_10235.map.gz | 49 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10235-v30.xml emd-10235-v30.xml emd-10235.xml emd-10235.xml | 11.4 KB 11.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10235_fsc.xml emd_10235_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_10235.png emd_10235.png | 241.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10235 http://ftp.pdbj.org/pub/emdb/structures/EMD-10235 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10235 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10235 | HTTPS FTP |
-Related structure data
| Related structure data |  6smhMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10235.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10235.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-electron microscopy structure of a RbcL8-Raf18 supercomplex from the cyanobacterium Synechococcus elongatus 7942 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.25 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : RbcL-Raf1 supercomplex of Synechococcus elongatus 7942
| Entire | Name: RbcL-Raf1 supercomplex of Synechococcus elongatus 7942 |
|---|---|
| Components |
|
-Supramolecule #1: RbcL-Raf1 supercomplex of Synechococcus elongatus 7942
| Supramolecule | Name: RbcL-Raf1 supercomplex of Synechococcus elongatus 7942 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: cryoEM structure of a RbcL8-Raf18 complex from Synechococcus elongatus 7942 |
|---|---|
| Source (natural) | Organism:  Synechococcus elongatus PCC 7942 (bacteria) Synechococcus elongatus PCC 7942 (bacteria) |
| Recombinant expression | Organism:  |
-Macromolecule #1: Ribulose bisphosphate carboxylase large chain
| Macromolecule | Name: Ribulose bisphosphate carboxylase large chain / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: ribulose-bisphosphate carboxylase |
|---|---|
| Source (natural) | Organism:  Synechococcus elongatus PCC 7942 = FACHB-805 (bacteria) Synechococcus elongatus PCC 7942 = FACHB-805 (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: LTYYTPDYTP KDTDLLAAFR FSPQPGVPAP EAGAAIAAES STGTWTTVWT DLLTDMDRYD GKCYHIEPVQ GEENSYFAFI ADPLDLFEE GSVTNILTSI VGNVFGFKAI RSLRLEDIRF PVALVKTFQG PPHGIQVERD LLNKYGRPML GCTIKPKLGL S AKNYGRAV ...String: LTYYTPDYTP KDTDLLAAFR FSPQPGVPAP EAGAAIAAES STGTWTTVWT DLLTDMDRYD GKCYHIEPVQ GEENSYFAFI ADPLDLFEE GSVTNILTSI VGNVFGFKAI RSLRLEDIRF PVALVKTFQG PPHGIQVERD LLNKYGRPML GCTIKPKLGL S AKNYGRAV YECLRGGLDF TKDDENINSQ PFQRWRDRFL FVADAIHKSQ AETGEIKGHY LNVTAPTCEE MMKRAEFAKE LG MPIIMHD FLTAGFTANT TLAKWCRDNG VLLHIHRAMH AVIDRQRNHG IHFRVLAKCL RLSGGDHLHS GTVVGKLEGD KAS TLGFVD LMREDHIEAD RSRGVFFTQD WASMPGVLPV ASGGIHVWHM PALVEIFGDD SVLQFGGGTL GHPWGNAPGA TANR VALEA CVQARNEGRD LYREGGDILR EAGKWSPELA AALDLWKEIK FE |
-Macromolecule #2: Rubisco accumulation factor 1 (RAF1)
| Macromolecule | Name: Rubisco accumulation factor 1 (RAF1) / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Synechococcus elongatus (strain PCC 7942 / FACHB-805) (bacteria) Synechococcus elongatus (strain PCC 7942 / FACHB-805) (bacteria) |
| Sequence | String: ERQELLGQLR RKEGRWLAWA RACQTLLKNG LNPQTLFEAT GFEPIQQNQI TVAMQVYDSI LRQDPPAHVR ETYQEWGSDL LYELRELDQ EQRSLCAQLA LERKLDADQI REVAKATKDF CRLPKQPENF DRHPGDAVAH QCWRLAQERT DLTERSRLIA R GLQFAQSA GARALIEALL LDLSGVPSRK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 20 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source: OTHER |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)