[English] 日本語
 Yorodumi
Yorodumi- EMDB-10017: Inward-open structure of the ASCT2 (SLC1A5) in absence of substrate -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10017 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Inward-open structure of the ASCT2 (SLC1A5) in absence of substrate | ||||||||||||||||||
 Map data Map data | None | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationglutamine secretion / L-glutamine import across plasma membrane / L-glutamine transmembrane transporter activity / L-serine transmembrane transporter activity / glutamine transport / ligand-gated channel activity / neutral amino acid transport / L-aspartate transmembrane transporter activity / L-aspartate import across plasma membrane / neutral L-amino acid transmembrane transporter activity ...glutamine secretion / L-glutamine import across plasma membrane / L-glutamine transmembrane transporter activity / L-serine transmembrane transporter activity / glutamine transport / ligand-gated channel activity / neutral amino acid transport / L-aspartate transmembrane transporter activity / L-aspartate import across plasma membrane / neutral L-amino acid transmembrane transporter activity / Amino acid transport across the plasma membrane / amino acid transmembrane transporter activity / symporter activity / antiporter activity / RHOJ GTPase cycle / RHOQ GTPase cycle / protein homotrimerization / amino acid transport / RHOH GTPase cycle / RAC3 GTPase cycle / transport across blood-brain barrier / RAC1 GTPase cycle / basal plasma membrane / erythrocyte differentiation / centriolar satellite / melanosome / signaling receptor activity / virus receptor activity / ciliary basal body / extracellular exosome / metal ion binding / membrane / plasma membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||||||||
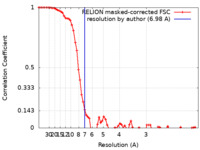
| Method | single particle reconstruction / cryo EM / Resolution: 6.98 Å | ||||||||||||||||||
 Authors Authors | Garaeva AA / Guskov A / Slotboom DJ / Paulino C | ||||||||||||||||||
| Funding support |  Netherlands, 5 items Netherlands, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: A one-gate elevator mechanism for the human neutral amino acid transporter ASCT2. Authors: Alisa A Garaeva / Albert Guskov / Dirk J Slotboom / Cristina Paulino /  Abstract: The human Alanine Serine Cysteine Transporter 2 (ASCT2) is a neutral amino acid exchanger that belongs to the solute carrier family 1 (SLC1A). SLC1A structures have revealed an elevator-type ...The human Alanine Serine Cysteine Transporter 2 (ASCT2) is a neutral amino acid exchanger that belongs to the solute carrier family 1 (SLC1A). SLC1A structures have revealed an elevator-type mechanism, in which the substrate is translocated across the cell membrane by a large displacement of the transport domain, whereas a small movement of hairpin 2 (HP2) gates the extracellular access to the substrate-binding site. However, it has remained unclear how substrate binding and release is gated on the cytoplasmic side. Here, we present an inward-open structure of the human ASCT2, revealing a hitherto elusive SLC1A conformation. Strikingly, the same structural element (HP2) serves as a gate in the inward-facing as in the outward-facing state. The structures reveal that SLC1A transporters work as one-gate elevators. Unassigned densities near the gate and surrounding the scaffold domain, may represent potential allosteric binding sites, which could guide the design of lipidic-inhibitors for anticancer therapy. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10017.map.gz emd_10017.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10017-v30.xml emd-10017-v30.xml emd-10017.xml emd-10017.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10017_fsc.xml emd_10017_fsc.xml | 7.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_10017.png emd_10017.png | 89.4 KB | ||
| Masks |  emd_10017_msk_1.map emd_10017_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Others |  emd_10017_half_map_1.map.gz emd_10017_half_map_1.map.gz emd_10017_half_map_2.map.gz emd_10017_half_map_2.map.gz | 23.3 MB 23.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10017 http://ftp.pdbj.org/pub/emdb/structures/EMD-10017 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10017 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10017 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10017.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10017.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.012 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
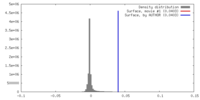
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10017_msk_1.map emd_10017_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
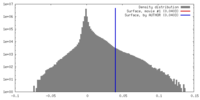
| Density Histograms |
-Half map: half map 2 used during refinement and for...
| File | emd_10017_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 used during refinement and for FSC gold-standard resolution calculation of apo ASCT2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
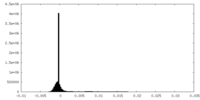
| Density Histograms |
-Half map: half map 1 used during refinement and for...
| File | emd_10017_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 used during refinement and for FSC gold-standard resolution calculation of apo ASCT2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ASCT2 (Alanine Serine Cysteine Transporter 2, SLC1A)
| Entire | Name: ASCT2 (Alanine Serine Cysteine Transporter 2, SLC1A) |
|---|---|
| Components |
|
-Supramolecule #1: ASCT2 (Alanine Serine Cysteine Transporter 2, SLC1A)
| Supramolecule | Name: ASCT2 (Alanine Serine Cysteine Transporter 2, SLC1A) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: in absence of substrate |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Molecular weight | Theoretical: 172 KDa |
-Macromolecule #1: ASCT2
| Macromolecule | Name: ASCT2 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MVADPPRDSK GLAAAEPTAN GGLALASIED QGAAAGGYCG SRDQVRRCLR ANLLVLLTVV AVVAGVALG LGVSGAGGAL ALGPERLSAF VFPGELLLRL LRMIILPLVV CSLIGGAASL D PGALGRLG AWALLFFLVT TLLASALGVG LALALQPGAA SAAINASVGA ...String: MVADPPRDSK GLAAAEPTAN GGLALASIED QGAAAGGYCG SRDQVRRCLR ANLLVLLTVV AVVAGVALG LGVSGAGGAL ALGPERLSAF VFPGELLLRL LRMIILPLVV CSLIGGAASL D PGALGRLG AWALLFFLVT TLLASALGVG LALALQPGAA SAAINASVGA AGSAENAPSK EV LDSFLDL ARNIFPSNLV SAAFRSYSTT YEERNITGTR VKVPVGQEVE GMNILGLVVF AIV FGVALR KLGPEGELLI RFFNSFNEAT MVLVSWIMWY APVGIMFLVA GKIVEMEDVG LLFA RLGKY ILCCLLGHAI HGLLVLPLIY FLFTRKNPYR FLWGIVTPLA TAFGTSSSSA TLPLM MKCV EENNGVAKHI SRFILPIGAT VNMDGAALFQ CVAAVFIAQL SQQSLDFVKI ITILVT ATA SSVGAAGIPA GGVLTLAIIL EAVNLPVDHI SLILAVDWLV DRSCTVLNVE GDALGAG LL QNYVDRTESR STEPELIQVK SELPLDPLPV PTEEGNPLLK HYRGPAGDAT VASEKESV M HHHHHH |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL |
|---|---|
| Buffer | pH: 7.4 Details: 20 mM Tris pH 7.4, 300 mM NaCl, 0.05% DDM, 0.005% CHS |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 288 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 90.0 K / Max: 105.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-60 / Number grids imaged: 6 / Number real images: 4305 / Average exposure time: 9.0 sec. / Average electron dose: 52.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 2.0 µm / Calibrated defocus min: 0.3 µm / Calibrated magnification: 49407 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.3 µm / Nominal magnification: 49407 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)