[English] 日本語
 Yorodumi
Yorodumi- EMDB-0995: Ultra-high voltage electron microscope tomography using 700-nm-th... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0995 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
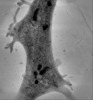
| Title | Ultra-high voltage electron microscope tomography using 700-nm-thick neurite section acquired at 15,000 magnification at an accelerating voltage of 1 MV | |||||||||
 Map data Map data | 700nm-thick neurite section of cultured PC12 cells (Magnification 15K) | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | electron tomography / negative staining | |||||||||
 Authors Authors | Nishida T / Yoshimura R / Nishi R / Imoto Y / Endo Y | |||||||||
 Citation Citation |  Journal: J Microsc / Year: 2020 Journal: J Microsc / Year: 2020Title: Application of ultra-high voltage electron microscope tomography to 3D imaging of microtubules in neurites of cultured PC12 cells. Authors: T Nishida / R Yoshimura / R Nishi / Y Imoto / Y Endo /  Abstract: Electron tomography methods using the conventional transmission electron microscope have been widely used to investigate the three-dimensional distribution patterns of various cellular structures ...Electron tomography methods using the conventional transmission electron microscope have been widely used to investigate the three-dimensional distribution patterns of various cellular structures including microtubules in neurites. Because the penetrating power of electrons depends on the section thickness and accelerating voltage, conventional TEM, having acceleration voltages up to 200 kV, is limited to sample thicknesses of 0.2 µm or less. In this paper, we show that the ultra-high voltage electron microscope (UHVEM), employing acceleration voltages of higher than 1000 kV (1 MV), allowed distinct reconstruction of the three-dimensional array of microtubules in a 0.7-µm-thick neurite section. The detailed structure of microtubules was more clearly reconstructed from a 0.7-µm-thick section at an accelerating voltage of 1 MV compared with a 1.0 µm section at 2 MV. Furthermore, the entire distribution of each microtubule in a neurite could be reconstructed from serial-section UHVEM tomography. Application of optimised UHVEM tomography will provide new insights, bridging the gap between the structure and function of widely-distributed cellular organelles such as microtubules for neurite outgrowth. LAY DESCRIPTION: An optimal 3D visualisation of microtubule cytoskeleton using ultra-high voltage electron microscopy tomography The ultra-high voltage electron microscope (UHVEM) is able to visualise a micrometre-thick specimen at nanoscale spatial resolution because of the high-energy electron beam penetrating such a specimen. In this study, we determined the optimal conditions necessary for microtubule cytoskeleton imaging within 0.7-µm-thick section using a combination with UHVEM and electron tomography method. Our approach provides excellent 3D information about the complex arrangement of the individual microtubule filaments that make up the microtubule network. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0995.map.gz emd_0995.map.gz | 127.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0995-v30.xml emd-0995-v30.xml emd-0995.xml emd-0995.xml | 8.6 KB 8.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0995.png emd_0995.png | 118.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0995 http://ftp.pdbj.org/pub/emdb/structures/EMD-0995 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0995 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0995 | HTTPS FTP |
-Related structure data
| Related structure data |  0987C  0994C  0996C C: citing same article ( |
|---|---|
| EM raw data |  EMPIAR-10355 (Title: Ultra-high voltage electron microscope tomography tilt series of 0.7-μm-thick neurite section acquired at 15,000× magnification at an accelerating voltage of 1 MV EMPIAR-10355 (Title: Ultra-high voltage electron microscope tomography tilt series of 0.7-μm-thick neurite section acquired at 15,000× magnification at an accelerating voltage of 1 MVData size: 120.3 MB Data #1: Ultra-high voltage electron microscope tomography tilt series of 0.7-μm-thick neurite section acquired at 15,000× magnification at an accelerating voltage of 1 MV [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0995.map.gz / Format: CCP4 / Size: 174.2 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) Download / File: emd_0995.map.gz / Format: CCP4 / Size: 174.2 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 700nm-thick neurite section of cultured PC12 cells (Magnification 15K) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 45.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Microtubule distribution in neurite of differentiated PC12 cell
| Entire | Name: Microtubule distribution in neurite of differentiated PC12 cell |
|---|---|
| Components |
|
-Supramolecule #1: Microtubule distribution in neurite of differentiated PC12 cell
| Supramolecule | Name: Microtubule distribution in neurite of differentiated PC12 cell type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.3 |
|---|---|
| Staining | Type: POSITIVE / Material: Uranyl Acetate, Lead |
| Sugar embedding | Material: resin |
| Grid | Material: COPPER / Mesh: 50 / Support film - Material: FORMVAR |
| Sectioning | Ultramicrotomy - Instrument: Reichert-Jung Ultracut E / Ultramicrotomy - Temperature: 300 K / Ultramicrotomy - Final thickness: 700 nm |
| Fiducial marker | Manufacturer: BBI Solutions / Diameter: 20 nm |
- Electron microscopy
Electron microscopy
| Microscope | HITACHI H3000 UHVEM |
|---|---|
| Image recording | Film or detector model: OTHER / Average electron dose: 13.0 e/Å2 |
| Electron beam | Acceleration voltage: 1000 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal magnification: 15000 |
| Sample stage | Specimen holder model: OTHER |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Software - Name:  IMOD / Number images used: 67 IMOD / Number images used: 67 |
|---|
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)