+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0789 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negatively stained reconstruction of a rubisco activase | |||||||||
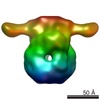
 Map data Map data | Negative stained 3D map of the rubisco activase | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Acidithiobacillus ferrooxidans (bacteria) Acidithiobacillus ferrooxidans (bacteria) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 16.0 Å | |||||||||
 Authors Authors | Tsai Y / Liu D / Bhushan S / Mueller-Cajar O | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2020 Journal: Proc Natl Acad Sci U S A / Year: 2020Title: Insights into the mechanism and regulation of the CbbQO-type Rubisco activase, a MoxR AAA+ ATPase. Authors: Yi-Chin Candace Tsai / Fuzhou Ye / Lynette Liew / Di Liu / Shashi Bhushan / Yong-Gui Gao / Oliver Mueller-Cajar /  Abstract: The vast majority of biological carbon dioxide fixation relies on the function of ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco). In most cases the enzyme exhibits a tendency to become ...The vast majority of biological carbon dioxide fixation relies on the function of ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco). In most cases the enzyme exhibits a tendency to become inhibited by its substrate RuBP and other sugar phosphates. The inhibition is counteracted by diverse molecular chaperones known as Rubisco activases (Rcas). In some chemoautotrophic bacteria, the CbbQO-type Rca Q2O2 repairs inhibited active sites of hexameric form II Rubisco. The 2.2-Å crystal structure of the MoxR AAA+ protein CbbQ2 from reveals the helix 2 insert (H2I) that is critical for Rca function and forms the axial pore of the CbbQ hexamer. Negative-stain electron microscopy shows that the essential CbbO adaptor protein binds to the conserved, concave side of the CbbQ2 hexamer. Site-directed mutagenesis supports a model in which adenosine 5'-triphosphate (ATP)-powered movements of the H2I are transmitted to CbbO via the concave residue L85. The basal ATPase activity of Q2O2 Rca is repressed but strongly stimulated by inhibited Rubisco. The characterization of multiple variants where this repression is released indicates that binding of inhibited Rubisco to the C-terminal CbbO VWA domain initiates a signal toward the CbbQ active site that is propagated via elements that include the CbbQ α4-β4 loop, pore loop 1, and the presensor 1-β hairpin (PS1-βH). Detailed mechanistic insights into the enzyme repair chaperones of the highly diverse CO fixation machinery of Proteobacteria will facilitate their successful implementation in synthetic biology ventures. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0789.map.gz emd_0789.map.gz | 364.8 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0789-v30.xml emd-0789-v30.xml emd-0789.xml emd-0789.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0789.png emd_0789.png | 28.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0789 http://ftp.pdbj.org/pub/emdb/structures/EMD-0789 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0789 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0789 | HTTPS FTP |
-Validation report
| Summary document |  emd_0789_validation.pdf.gz emd_0789_validation.pdf.gz | 78.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0789_full_validation.pdf.gz emd_0789_full_validation.pdf.gz | 77.5 KB | Display | |
| Data in XML |  emd_0789_validation.xml.gz emd_0789_validation.xml.gz | 495 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0789 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0789 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0789 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0789 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0789.map.gz / Format: CCP4 / Size: 489.3 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0789.map.gz / Format: CCP4 / Size: 489.3 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stained 3D map of the rubisco activase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.19 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : CBBQO type Rubisco activase
| Entire | Name: CBBQO type Rubisco activase |
|---|---|
| Components |
|
-Supramolecule #1: CBBQO type Rubisco activase
| Supramolecule | Name: CBBQO type Rubisco activase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Acidithiobacillus ferrooxidans (bacteria) Acidithiobacillus ferrooxidans (bacteria) |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 0.27 kDa/nm |
-Macromolecule #1: Rubisco activase
| Macromolecule | Name: Rubisco activase / type: protein_or_peptide / ID: 1 / Enantiomer: DEXTRO |
|---|---|
| Source (natural) | Organism:  Acidithiobacillus ferrooxidans (bacteria) Acidithiobacillus ferrooxidans (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MTATDSSILN QYLVGKEPFY QPQHDEVALF EAAYRKRLPV MVKGPTGCGK SRFVEFMAWR LGKPLVTVAC NEDMTAADLV GRWLLDKDGT RWQDGPLTVA ARYGAICYLD EIVEARQDTT VVIHPLTDHR RTLPLDKKGE LIRAHPDFQL VISYNPGYQS LMKDLKQSTK ...String: MTATDSSILN QYLVGKEPFY QPQHDEVALF EAAYRKRLPV MVKGPTGCGK SRFVEFMAWR LGKPLVTVAC NEDMTAADLV GRWLLDKDGT RWQDGPLTVA ARYGAICYLD EIVEARQDTT VVIHPLTDHR RTLPLDKKGE LIRAHPDFQL VISYNPGYQS LMKDLKQSTK QRFTGFEFDY PNAELEAGIL VQETGVAPSI AAQLVTVAAT ARRLKGHGLD EGISTRLLVY AAMLMDDGVA PRAACRMALV QPITDDADIR ATLEHAIDMT FA MTDHPRD ASLPSRLAAY RKQLDCRFPR VGEVFSDCIA KASARLGPAG VSAYVDAARA LCKLGRGEEP VLIFLEEWPD VGAALGDGTL EMVMQMVQFM QRTPNGNAIG GFLQSLAPVS RALLSREQLG HYLNTLRDMM ERTTGSIHGH HQTHPSPGLP ELLRQAPTLL QSLTVDGLRN WVDYGVRNYL HHPERQKDFF SLQSADSRAV LQRERHGTLL ADVERKLDLY LRGLWQDSEV LVPYSTAFAT LRTPQPYYDA LGMRLPDVLD DLPGIGALDH YRAILAHMVG HRRWSTPQIA DNWSPFQRLA VEFFEDARID TLLIRTYPGL RTLFLALHPK PGEDACDPET TSCLRHRLAM LSRALLDPEH GYRNPVLHDF VDRFHGELAG GNADTATMAR LALDYVTTTR RQSDQFAKVH FADTEISYRD DNRGLWRFIE SGDEEEAFDA EQRKAPDLET QGLPPRHYPE WDYQSQSYRP DWVSLYEGLH ASAPAATIDQ LLLKHAALAK HLKRLLDLLK PQDKVRIRYQ EEGSELDLDV ALRSWIDFKS GSTPDPRINM SHRTAGRDIA VTLLLDLSES LNESVKTGGG DGQTVLQLSQ EAVSLLAWSI EQLGDPLAIA GFNSNTRHEV RYQHIKGFSE PWGDVVKGRL AALQAGYSTR MGAAMRHAGH YLATRKADKK LMLVLTDGRP SDVDVQDDRL LIEDARQAVN ELDRDGIFTY CISLDPHADA YVADIFGRQY TVIDHIARLP EKLPELFIAL TR |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.05 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20 mM Tris-HCl pH8.0, 50 mM NaCl |
| Staining | Type: NEGATIVE / Material: Uranyl Acetate Details: The sample was applied to a carbon-coated TEM grid and stained with 2% (w/v) uranyl acetate |
| Grid | Model: Homemade / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 6.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Details | Monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI 12 |
|---|---|
| Image recording | Film or detector model: FEI EAGLE (4k x 4k) / Number grids imaged: 1 / Number real images: 60 / Average exposure time: 1.0 sec. / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Calibrated defocus max: 1.5 µm / Calibrated defocus min: 1.25 µm / Calibrated magnification: 49000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 6.3 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.25 µm |
| Sample stage | Specimen holder model: GATAN 910 MULTI-SPECIMEN SINGLE TILT CRYO TRANSFER HOLDER |
 Movie
Movie Controller
Controller



 UCSF Chimera
UCSF Chimera






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















