[English] 日本語
 Yorodumi
Yorodumi- EMDB-0713: Complex structure of Iota toxin enzymatic component (Ia) and bind... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0713 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with short stem | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bacterial binary toxin / Protein translocation channel / TOXIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Yoshida T / Yamada T | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2020 Journal: Nat Struct Mol Biol / Year: 2020Title: Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex. Authors: Tomohito Yamada / Toru Yoshida / Akihiro Kawamoto / Kaoru Mitsuoka / Kenji Iwasaki / Hideaki Tsuge /  Abstract: The iota toxin produced by Clostridium perfringens type E is a binary toxin comprising two independent polypeptides: Ia, an ADP-ribosyltransferase, and Ib, which is involved in cell binding and ...The iota toxin produced by Clostridium perfringens type E is a binary toxin comprising two independent polypeptides: Ia, an ADP-ribosyltransferase, and Ib, which is involved in cell binding and translocation of Ia across the cell membrane. Here we report cryo-EM structures of the translocation channel Ib-pore and its complex with Ia. The high-resolution Ib-pore structure demonstrates a similar structural framework to that of the catalytic ϕ-clamp of the anthrax protective antigen pore. However, the Ia-bound Ib-pore structure shows a unique binding mode of Ia: one Ia binds to the Ib-pore, and the Ia amino-terminal domain forms multiple weak interactions with two additional Ib-pore constriction sites. Furthermore, Ib-binding induces tilting and partial unfolding of the Ia N-terminal α-helix, permitting its extension to the ϕ-clamp gate. This new mechanism of N-terminal unfolding is crucial for protein translocation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0713.map.gz emd_0713.map.gz | 10.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0713-v30.xml emd-0713-v30.xml emd-0713.xml emd-0713.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
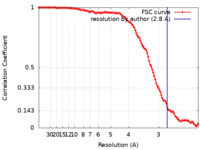
| FSC (resolution estimation) |  emd_0713_fsc.xml emd_0713_fsc.xml | 11.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0713.png emd_0713.png | 137.7 KB | ||
| Filedesc metadata |  emd-0713.cif.gz emd-0713.cif.gz | 6.6 KB | ||
| Others |  emd_0713_additional.map.gz emd_0713_additional.map.gz emd_0713_additional_1.map.gz emd_0713_additional_1.map.gz | 98.1 MB 98.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0713 http://ftp.pdbj.org/pub/emdb/structures/EMD-0713 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0713 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0713 | HTTPS FTP |
-Related structure data
| Related structure data |  6kloMC  0720C  0721C  6klwC  6klxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0713.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0713.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.13 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Postprocessing is not applied.
| File | emd_0713_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessing is not applied. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Postprocessing is not applied.
| File | emd_0713_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Postprocessing is not applied. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Complex structure of Iota toxin enzymatic component (Ia) and bind...
| Entire | Name: Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with short stem |
|---|---|
| Components |
|
-Supramolecule #1: Complex structure of Iota toxin enzymatic component (Ia) and bind...
| Supramolecule | Name: Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with short stem type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 570 KDa |
-Macromolecule #1: Iota toxin component Ib
| Macromolecule | Name: Iota toxin component Ib / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 74.37882 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SAAWEDEDLD TDNDNIPDAY EKNGYTIKDS IAVKWNDSFA EQGYKKYVSS YLESNTAGDP YTDYQKASGS IDKAIKLEAR DPLVAAYPV VGVGMENLII STNEHASSDQ GKTVSRATTN SKTDANTVGV SISAGYQNGF TGNITTSYSH TTDNSTAVQD S NGESWNTG ...String: SAAWEDEDLD TDNDNIPDAY EKNGYTIKDS IAVKWNDSFA EQGYKKYVSS YLESNTAGDP YTDYQKASGS IDKAIKLEAR DPLVAAYPV VGVGMENLII STNEHASSDQ GKTVSRATTN SKTDANTVGV SISAGYQNGF TGNITTSYSH TTDNSTAVQD S NGESWNTG LSINKGESAY INANVRYYNT GTAPMYKVTP TTNLVLDGET LATIKAQDNQ IGNNLSPNET YPKKGLSPLA LN TMDQFNA RLIPINYDQL KKLDSGKQIK LETTQVSGNY GTKNSQGQII TEGNSWSNYI SQIDSVSASI ILDTGSQTFE RRV AAKEQG NPEDKTPEIT IGEAIKKAFS ATKNGELLYF NGIPIDESCV ELIFDDNTSE IIKEQLKYLD DKKIYNVKLE RGMN ILIKV PSYFTNFDEY NNFPASWSNI DTKNQDGLQS VANKLSGETK IIIPMSKLKP YKRYVFSGYS KDPSTSNSIT VNIKS KEQK TDYLVPEKDY TKFSYEFETT GKDSSDIEIT LTSSGVIFLD NLSITELNST PEILKEPEIK VPSDQEILDA HNKYYA DIK LDTNTGNTYI DGIYFEPTQT NKEALDYIQK YRVEATLQYS GFKDIGTKDK EIRNYLGDQN QPKTNYINFR SYFTSGE NV MTYKKLRIYA VTPDNRELLV LSVN UniProtKB: Iota toxin component Ib |
-Macromolecule #2: Iota toxin component Ia
| Macromolecule | Name: Iota toxin component Ia / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.061988 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSHMAFIERP EDFLKDKENA IQWEKKEAER VEKNLDTLEK EALELYKKDS EQISNYSQTR QYFYDYQIES NPREKEYKNL RNAISKNKI DKPINVYYFE SPEKFAFNKE IRTENQNEIS LEKFNELKET IQDKLFKQDG FKDVSLYEPG NGDEKPTPLL I HLKLPKNT ...String: GSHMAFIERP EDFLKDKENA IQWEKKEAER VEKNLDTLEK EALELYKKDS EQISNYSQTR QYFYDYQIES NPREKEYKNL RNAISKNKI DKPINVYYFE SPEKFAFNKE IRTENQNEIS LEKFNELKET IQDKLFKQDG FKDVSLYEPG NGDEKPTPLL I HLKLPKNT GMLPYINSND VKTLIEQDYS IKIDKIVRIV IEGKQYIKAE ASIVNSLDFK DDVSKGDLWG KENYSDWSNK LT PNELADV NDYMRGGYTA INNYLISNGP LNNPNPELDS KVNNIENALK LTPIPSNLIV YRRSGPQEFG LTLTSPEYDF NKI ENIDAF KEKWEGKVIT YPNFISTSIG SVNMSAFAKR KIILRINIPK DSPGAYLSAI PGYAGEYEVL LNHGSKFKIN KVDS YKDGT VTKLILDATL IN UniProtKB: Iota toxin component Ia |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 14 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.48 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 78.9 K / Max: 78.9 K |
| Specialist optics | Spherical aberration corrector: Microscope was modified with a Cs corrector |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 2151 / Average exposure time: 84.09 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)