[English] 日本語
 Yorodumi
Yorodumi- EMDB-0499: CryoEM structure of Helicobacter pylori urea channel in open state. -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0499 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Helicobacter pylori urea channel in open state. | |||||||||
 Map data Map data | em-volume_P1.map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Helicobacter pylori / urea channel / open state / TRANSPORT PROTEIN | |||||||||
| Function / homology | AmiS/UreI transporter / AmiS/UreI transporter superfamily / AmiS/UreI family transporter / identical protein binding / plasma membrane / Acid-activated urea channel Function and homology information Function and homology information | |||||||||
| Biological species |   Helicobacter pylori (strain J99 / ATCC 700824) (bacteria) Helicobacter pylori (strain J99 / ATCC 700824) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Cui YX / Zhou K | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2019 Journal: Sci Adv / Year: 2019Title: pH-dependent gating mechanism of the urea channel revealed by cryo-EM. Authors: Yanxiang Cui / Kang Zhou / David Strugatsky / Yi Wen / George Sachs / Z Hong Zhou / Keith Munson /  Abstract: The urea channel of (UreI) is an ideal drug target for preventing gastric cancer but incomplete understanding of its gating mechanism has hampered development of inhibitors for the eradication of . ...The urea channel of (UreI) is an ideal drug target for preventing gastric cancer but incomplete understanding of its gating mechanism has hampered development of inhibitors for the eradication of . Here, we present the cryo-EM structures of UreI in closed and open conformations, both at a resolution of 2.7 Å. Our hexameric structures of this small membrane protein (~21 kDa/protomer) resolve its periplasmic loops and carboxyl terminus that close and open the channel, and define a gating mechanism that is pH dependent and requires cooperativity between protomers in the hexamer. Gating is further associated with well-resolved changes in the channel-lining residues that modify the shape and length of the urea pore. Site-specific mutations in the periplasmic domain and urea pore identified key residues important for channel function. Drugs blocking the urea pore based on our structures should lead to a new strategy for eradication. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0499.map.gz emd_0499.map.gz | 14.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0499-v30.xml emd-0499-v30.xml emd-0499.xml emd-0499.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0499_fsc.xml emd_0499_fsc.xml | 5.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_0499.png emd_0499.png | 3 MB | ||
| Masks |  emd_0499_msk_1.map emd_0499_msk_1.map | 15.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-0499.cif.gz emd-0499.cif.gz | 6.3 KB | ||
| Others |  emd_0499_half_map_1.map.gz emd_0499_half_map_1.map.gz emd_0499_half_map_2.map.gz emd_0499_half_map_2.map.gz | 11.1 MB 11.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0499 http://ftp.pdbj.org/pub/emdb/structures/EMD-0499 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0499 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0499 | HTTPS FTP |
-Validation report
| Summary document |  emd_0499_validation.pdf.gz emd_0499_validation.pdf.gz | 813.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0499_full_validation.pdf.gz emd_0499_full_validation.pdf.gz | 812.7 KB | Display | |
| Data in XML |  emd_0499_validation.xml.gz emd_0499_validation.xml.gz | 11.4 KB | Display | |
| Data in CIF |  emd_0499_validation.cif.gz emd_0499_validation.cif.gz | 15.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0499 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0499 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0499 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0499 | HTTPS FTP |
-Related structure data
| Related structure data |  6nskMC  0498C  6nsjC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0499.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0499.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | em-volume_P1.map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
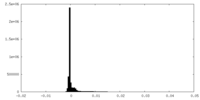
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0499_msk_1.map emd_0499_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
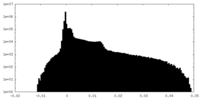
| Density Histograms |
-Half map: em-half-volume P1
| File | emd_0499_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | em-half-volume_P1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: em-half-volume P2
| File | emd_0499_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | em-half-volume_P2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : urea channel of Helicobacter pylori
| Entire | Name: urea channel of Helicobacter pylori |
|---|---|
| Components |
|
-Supramolecule #1: urea channel of Helicobacter pylori
| Supramolecule | Name: urea channel of Helicobacter pylori / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 20 KDa |
-Macromolecule #1: Acid-activated urea channel
| Macromolecule | Name: Acid-activated urea channel / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Helicobacter pylori (strain J99 / ATCC 700824) (bacteria) Helicobacter pylori (strain J99 / ATCC 700824) (bacteria)Strain: J99 / ATCC 700824 |
| Molecular weight | Theoretical: 22.533258 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLGLVLLYVG IVLISNGICG LTKVDPKSTA VMNFFVGGLS IVCNVVVITY SALHPTAPVE GHHHHHHAED IVQVSHHLTS FYGPATGLL FGFTYLYAAI NHTFGLDWRP YSWYSLFVAI NTVPAAILSH YSDMLDDHKV LGITEGDWWA IIWLAWGVLW L TAFIENIL ...String: MLGLVLLYVG IVLISNGICG LTKVDPKSTA VMNFFVGGLS IVCNVVVITY SALHPTAPVE GHHHHHHAED IVQVSHHLTS FYGPATGLL FGFTYLYAAI NHTFGLDWRP YSWYSLFVAI NTVPAAILSH YSDMLDDHKV LGITEGDWWA IIWLAWGVLW L TAFIENIL KIPLGKFTPW LAIIEGILTA WIPAWLLFIQ HWV UniProtKB: Acid-activated urea channel |
-Macromolecule #2: 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE
| Macromolecule | Name: 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE / type: ligand / ID: 2 / Number of copies: 18 / Formula: XP4 |
|---|---|
| Molecular weight | Theoretical: 591.777 Da |
| Chemical component information |  ChemComp-XP4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| ||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: FORMVAR / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 98.0 K / Max: 100.0 K |
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 2-59 / Number grids imaged: 1 / Number real images: 2901 / Average exposure time: 9.0 sec. / Average electron dose: 43.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 2.6 µm / Calibrated defocus min: 1.2 µm / Calibrated magnification: 46730 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.4000000000000001 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)