+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0204 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | OBP chaperonin in the nucleotide-free state | |||||||||
 Map data Map data | A symmetry-free cryo-EM structure of a single-ring chaperonin encoded by gene 246 of bacteriophage OBP Pseudomonas fluorescens in the nucleotide-free state. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | chaperonin / nucleotide-free / CHAPERONE | |||||||||
| Function / homology |  Function and homology information Function and homology informationATP-dependent protein folding chaperone / protein refolding / ATP binding Similarity search - Function | |||||||||
| Biological species |  Pseudomonas phage OBP (virus) Pseudomonas phage OBP (virus) | |||||||||
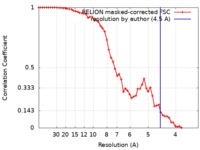
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Stanishneva-Konovalova TB / Pichkur EB / Sokolova OS | |||||||||
| Funding support |  Russian Federation, 2 items Russian Federation, 2 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2020 Journal: J Struct Biol / Year: 2020Title: Cryo-EM reveals an asymmetry in a novel single-ring viral chaperonin. Authors: Tatiana B Stanishneva-Konovalova / Pavel I Semenyuk / Lidia P Kurochkina / Evgeny B Pichkur / Alexander L Vasilyev / Mikhail V Kovalchuk / Mikhail P Kirpichnikov / Olga S Sokolova /  Abstract: Chaperonins are ubiquitously present protein complexes, which assist the proper folding of newly synthesized proteins and prevent aggregation of denatured proteins in an ATP-dependent manner. They ...Chaperonins are ubiquitously present protein complexes, which assist the proper folding of newly synthesized proteins and prevent aggregation of denatured proteins in an ATP-dependent manner. They are classified into group I (bacterial, mitochondrial, chloroplast chaperonins) and group II (archaeal and eukaryotic cytosolic variants). However, both of these groups do not include recently discovered viral chaperonins. Here, we solved the symmetry-free cryo-EM structures of a single-ring chaperonin encoded by the gene 246 of bacteriophage OBP Pseudomonas fluorescens, in the nucleotide-free, ATPγS-, and ADP-bound states, with resolutions of 4.3 Å, 5.0 Å, and 6 Å, respectively. The structure of OBP chaperonin reveals a unique subunit arrangement, with three pairs of subunits and one unpaired subunit. Each pair combines subunits in two possible conformations, differing in nucleotide-binding affinity. The binding of nucleotides results in the increase of subunits' conformational variability. Due to its unique structural and functional features, OBP chaperonin can represent a new group. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0204.map.gz emd_0204.map.gz | 56.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0204-v30.xml emd-0204-v30.xml emd-0204.xml emd-0204.xml | 12.7 KB 12.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0204_fsc.xml emd_0204_fsc.xml | 5.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_0204.png emd_0204.png | 203.7 KB | ||
| Filedesc metadata |  emd-0204.cif.gz emd-0204.cif.gz | 5.3 KB | ||
| Others |  emd_0204_additional.map.gz emd_0204_additional.map.gz | 9.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0204 http://ftp.pdbj.org/pub/emdb/structures/EMD-0204 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0204 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0204 | HTTPS FTP |
-Related structure data
| Related structure data |  6hddMC  0191C  0208C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0204.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0204.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A symmetry-free cryo-EM structure of a single-ring chaperonin encoded by gene 246 of bacteriophage OBP Pseudomonas fluorescens in the nucleotide-free state. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: A symmetry-free cryo-EM structure of a single-ring chaperonin...
| File | emd_0204_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A symmetry-free cryo-EM structure of a single-ring chaperonin encoded by gene 246 of bacteriophage OBP Pseudomonas fluorescens in the nucleotide-free state. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : OBP chaperonin in the nucleotide-free state
| Entire | Name: OBP chaperonin in the nucleotide-free state |
|---|---|
| Components |
|
-Supramolecule #1: OBP chaperonin in the nucleotide-free state
| Supramolecule | Name: OBP chaperonin in the nucleotide-free state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage OBP (virus) Pseudomonas phage OBP (virus) |
| Molecular weight | Theoretical: 420 KDa |
-Macromolecule #1: Putative chaperonin GroEL
| Macromolecule | Name: Putative chaperonin GroEL / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage OBP (virus) Pseudomonas phage OBP (virus) |
| Molecular weight | Theoretical: 57.259703 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TEISFTNKND TREIVDEVVE NAFSSVCSTM GPNGNYVVIN QLNSPKVTKD GVSVARALDF NEARRNMIAK IITEPSIKTD AEVGDGTTT TVFITYHLYQ KFKDAMSFAN TRYLDTLIKQ VLQYIGTLIQ PGEIESEMFR NMLLTSSNYE EEIVDKILDI Y REHKNPNI ...String: TEISFTNKND TREIVDEVVE NAFSSVCSTM GPNGNYVVIN QLNSPKVTKD GVSVARALDF NEARRNMIAK IITEPSIKTD AEVGDGTTT TVFITYHLYQ KFKDAMSFAN TRYLDTLIKQ VLQYIGTLIQ PGEIESEMFR NMLLTSSNYE EEIVDKILDI Y REHKNPNI HLEKSPMLPA DEVKMTKEIY FEGSFPIETQ VPANGAYVVG PEKVGVVLID GSIRAYPTQL INALLNRFID NP VVLMARN FEPEVIAAIN NENQRLGTSR IFAYKVNAAG LLGAGTIDDL GRLLNIGPVF DVNSVDPALV KYNDVTLWLG RKG ILLDKS IEEVESRADS ILEGLDNRYE ALGIIERQTP IGRELNRRIG RLRANNVTIK VTGVTVSDAS ERWARYEDVM KAAR TGQQF GVIPGIGYGY LMASKWLEAN VPQQSDEKLE KCRIGLIEVL RAQYEHLTGH DGSAENPIFI DLVTGQESDT PMNVY DNAA ATMIALEGAW QTAKTLGKIS NVMGRSNTNY A UniProtKB: Putative chaperonin GroEL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 100.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)