[English] 日本語
 Yorodumi
Yorodumi- EMDB-0093: Structure of human HCN4 hyperpolarization-activated cyclic nucleo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0093 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel | |||||||||
 Map data Map data | Cryosparc sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ION CHANNEL / PACEMAKER CURRENT / Structural Genomics / Structural Genomics Consortium / SGC / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationvoltage-gated potassium channel activity involved in SA node cell action potential depolarization / sinoatrial node development / HCN channels / regulation of cardiac muscle cell action potential involved in regulation of contraction / SA node cell action potential / membrane depolarization during SA node cell action potential / HCN channel complex / cellular response to cGMP / intracellularly cAMP-activated cation channel activity / regulation of SA node cell action potential ...voltage-gated potassium channel activity involved in SA node cell action potential depolarization / sinoatrial node development / HCN channels / regulation of cardiac muscle cell action potential involved in regulation of contraction / SA node cell action potential / membrane depolarization during SA node cell action potential / HCN channel complex / cellular response to cGMP / intracellularly cAMP-activated cation channel activity / regulation of SA node cell action potential / regulation of membrane depolarization / membrane depolarization during cardiac muscle cell action potential / sodium ion import across plasma membrane / blood circulation / voltage-gated sodium channel activity / potassium ion import across plasma membrane / regulation of heart rate by cardiac conduction / voltage-gated potassium channel activity / monoatomic cation transport / regulation of cardiac muscle contraction / cAMP binding / potassium ion transmembrane transport / regulation of heart rate / sodium ion transmembrane transport / muscle contraction / cellular response to cAMP / regulation of membrane potential / axon / dendrite / perinuclear region of cytoplasm / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
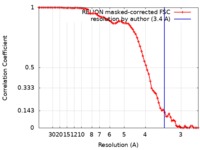
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Shintre CA / Pike ACW | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel Authors: Shintre CA / Pike ACW / Tessitore A / Young M / Bushell SR / Strain-Damerell C / Mukhopadhyay S / Burgess-Brown NA / Huiskonen JT / Arrowsmith CH / Edwards AM / Bountra C / Carpenter EP | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0093.map.gz emd_0093.map.gz | 38 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0093-v30.xml emd-0093-v30.xml emd-0093.xml emd-0093.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0093_fsc.xml emd_0093_fsc.xml | 7.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_0093.png emd_0093.png | 189.4 KB | ||
| Masks |  emd_0093_msk_1.map emd_0093_msk_1.map | 40.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-0093.cif.gz emd-0093.cif.gz | 6.5 KB | ||
| Others |  emd_0093_additional.map.gz emd_0093_additional.map.gz emd_0093_half_map_1.map.gz emd_0093_half_map_1.map.gz emd_0093_half_map_2.map.gz emd_0093_half_map_2.map.gz | 20.3 MB 37.2 MB 37.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0093 http://ftp.pdbj.org/pub/emdb/structures/EMD-0093 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0093 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0093 | HTTPS FTP |
-Related structure data
| Related structure data |  6gynMC  0094C  6gyoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0093.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0093.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0093_msk_1.map emd_0093_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
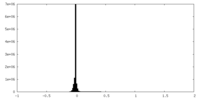
| Density Histograms |
-Additional map: Cryosparc unsharpened map
| File | emd_0093_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryosparc unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
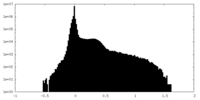
| Density Histograms |
-Half map: Half map1
| File | emd_0093_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
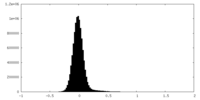
| Density Histograms |
-Half map: Half map2
| File | emd_0093_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
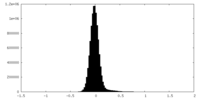
| Density Histograms |
- Sample components
Sample components
-Entire : Potassium/Sodium hyperpolarization-activated cyclic nucleotide-ga...
| Entire | Name: Potassium/Sodium hyperpolarization-activated cyclic nucleotide-gated ion channel 4 |
|---|---|
| Components |
|
-Supramolecule #1: Potassium/Sodium hyperpolarization-activated cyclic nucleotide-ga...
| Supramolecule | Name: Potassium/Sodium hyperpolarization-activated cyclic nucleotide-gated ion channel 4 type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Potassium/sodium hyperpolarization-activated cyclic nucleotide-ga...
| Macromolecule | Name: Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 60.836754 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SMLPEAEVRL GQAGFMQRQF GAMLQPGVNK FSLRMFGSQK AVEREQERVK SAGFWIIHPY SDFRFYWDLT MLLLMVGNLI IIPVGITFF KDENTTPWIV FNVVSDTFFL IDLVLNFRTG IVVEDNTEII LDPQRIKMKY LKSWFMVDFI SSIPVDYIFL I VETRIDSE ...String: SMLPEAEVRL GQAGFMQRQF GAMLQPGVNK FSLRMFGSQK AVEREQERVK SAGFWIIHPY SDFRFYWDLT MLLLMVGNLI IIPVGITFF KDENTTPWIV FNVVSDTFFL IDLVLNFRTG IVVEDNTEII LDPQRIKMKY LKSWFMVDFI SSIPVDYIFL I VETRIDSE VYKTARALRI VRFTKILSLL RLLRLSRLIR YIHQWEEIFH MTYDLASAVV RIVNLIGMML LLCHWDGCLQ FL VPMLQDF PDDCWVSINN MVNNSWGKQY SYALFKAMSH MLCIGYGRQA PVGMSDVWLT MLSMIVGATC YAMFIGHATA LIQ SLDSSR RQYQEKYKQV EQYMSFHKLP PDTRQRIHDY YEHRYQGKMF DEESILGELS EPLREEIINF NCRKLVASMP LFAN ADPNF VTSMLTKLRF EVFQPGDYII REGTIGKKMY FIQHGVVSVL TKGNKETKLA DGSYFGEICL LTRGRRTASV RADTY CRLY SLSVDNFNEV LEEYPMMRRA FETVALDRLD RIGKKNS UniProtKB: Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 |
-Macromolecule #2: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 2 / Number of copies: 28 / Formula: PC1 |
|---|---|
| Molecular weight | Theoretical: 790.145 Da |
| Chemical component information |  ChemComp-PC1: |
-Macromolecule #3: 1,2-Distearoyl-sn-glycerophosphoethanolamine
| Macromolecule | Name: 1,2-Distearoyl-sn-glycerophosphoethanolamine / type: ligand / ID: 3 / Number of copies: 8 / Formula: 3PE |
|---|---|
| Molecular weight | Theoretical: 748.065 Da |
| Chemical component information |  ChemComp-3PE: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV / Details: blotted for 5.5s before plunge. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Details | EBIC TITAN KRIOS M02 |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-48 / Number grids imaged: 1 / Number real images: 1294 / Average exposure time: 12.0 sec. / Average electron dose: 48.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.3 µm / Nominal magnification: 37313 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Model refined against the cryosparc b-factor sharpened map using default restraints |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-6gyn: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)